A complex containing lysine-acetylated actin inhibits the formin INF2
Preprint posted on 31 December 2018 https://www.biorxiv.org/content/10.1101/509133v1
Article now published in Nature Cell Biology at http://dx.doi.org/10.1038/s41556-019-0307-4
CAPing INF2 activity: Identification of a cellular factor that facilitates INF2 auto-inhibition
Selected by Laura McCormickCategories: biochemistry, cell biology
Background:
Actin filaments perform an amazingly diverse number of roles in the cell—ranging from cell division to vesicle trafficking to mitochondrial division. Underlying the distinct roles that different populations of polymerized actin can play are the myriad of proteins that regulate actin polymerization.
The formin family of proteins is united by their ability to nucleate actin filaments, overcoming the rate-limiting step of actin polymerization. In recent years, the Higgs lab has extensively studied inverted formin 2 (INF2). In addition to nucleating new actin filaments, INF2 enhances the elongation of actin filaments and accelerates actin depolymerization by severing filaments. Among other roles in the cell, INF2 regulates actin polymerization required for ER-mitochondria calcium transport and mitochondrial division.
A subset of formin proteins are regulated through autoinhibition between the conserved Diaphanous Inhibitory Domain (DID) and the C-terminal Diaphanous Auto-regulatory Domain (DAD). While cellular assays show that regulation of INF2 activity requires the DID-DAD interaction, purified INF2 protein is constitutively active in biochemical assays. To reconcile this disparity, the Higgs lab hypothesized that cellular factor enhances INF2 autoinhibition via the DID-DAD and properly regulates INF2 activity.
Key Findings:
To identify the INF2 inhibitor, the Higgs lab utilized a classic biochemical approach. Mouse brain cytosol was run through various column chromatography steps and fractionated. Each fraction was then mixed with purified INF2 protein to evaluate INF2’s ability to polymerize actin. Fractions that inhibited INF2 activity were pooled and continued through subsequent fractionation until the final Brain Inhibitory Fraction (BIF) was purified. While the BIF inhibited full length INF2 activity, it did not inhibit an INF2 construct lacking the DID domain, suggesting the BIF enhanced INF2 inhibition through DID-DAD interactions.
When the BIF was separated on a SDS-PAGE gel, five protein bands were visualized and sent for mass spectrometry identification. Notably, cyclase associated protein 1 and 2 (CAP1, CAP2) were identified. CAP proteins can bind actin monomers and enhance polymerization, as well as bind actin filaments and enhance severing. While immunodepletion of CAP2 eliminated the strong inhibitory action of the BIF, the authors found recombinant CAP (purified from HEK293 cells) only weakly inhibited INF2 activity in vitro.
Although the differences in inhibitory activity between brain purified and recombinant CAP were puzzling, the authors hypothesized that an addition protein may enhance INF2 inhibition. As CAP binds and co-purifies with actin monomers, they questioned if brain actin vs. HEK293 actin could affect the CAP-INF2 interaction. Following actin-exchange assays, the authors confirmed that recombinant CAP bound to brain actin inhibits INF2 significantly better than CAP bound to HEK293 actin. As most in vitro actin assays utilize actin purified from rabbit or chicken skeletal muscle, the authors also tested the inhibitory action of CAP exchanged with these actin sources. While CAP-chicken actin showed inhibitory activity comparable to CAP-brain actin, CAP-rabbit actin had minimal impact on INF2. To help interpret these disparities in inhibition between actin sources, they probed post-translational modifications of chicken actin by mass spec and identified 4 acetylated residues. Deacetylation of chicken actin by Histone Deacetylase 6 (HDAC6) significantly decreased the inhibitory effect of CAP-chicken actin on INF2 activity, confirming that actin lysine acetylation is key component of CAP-mediated INF2 inhibition.
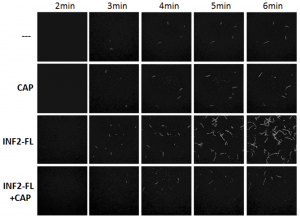
Fig. 1. TIRF microscopy of actin polymerization in vitro. While full-length INF2 (INF2-FL) alone enhances actin polymerization, the presence of CAP-actin strongly inhibits INF2 activity.
To understand the physiological role of CAP-acetylated actin inhibition and HDAC6-mediated deacetylation, the authors completed several cellular assays. Previous work in several labs, including the Higgs lab, has shown that ionomycin-induced calcium influx in cultured U2OS cells stimulates an INF2-mediated burst of actin polymerization and subsequent mitochondrial division. Interestingly, they found that treatment with Tubastatin A, an inhibitor of HDAC6, blocked the ionomycin-induced actin burst, suggesting that deacetylation of the CAP-actin complex is required for INF2 activation in cells. Supporting this hypothesis, they also found ionomycin treatment transiently decreases the amount of acetylated actin bound to CAP2, as well as the amount of INF2 co-purified with CAP. Interestingly, mutations in the INF2 DID domain have been observed in Charcot-Marie Tooth Disease and Focal Segmental Glomerulosclerosis. Cells transfected with INF2 constructs containing disease-linked mutations exhibit elevated levels of polymerized actin, suggesting these mutations increase basal INF2 activity. Furthermore, these INF2 disease constructs did not co-purify with CAP2 from U2OS cells and were weakly inhibited by CAP-actin in actin pyrene assays. Collectively, these data show that the CAP-acetylated actin complex facilitates INF2 autoinhibition and dysregulation of INF2 activity is linked to disease states.
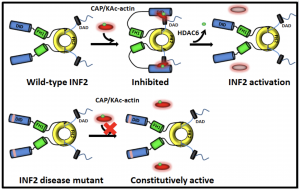
Fig 2. INF2 auto-inhibition is facilitated by the CAP-acetylated actin complex and is relieved by HDAC-6 mediated deacetylation of actin.
Why I Selected:
- In this paper, the authors employed classic column chromatography—old-school biochemistry—to identify the cellular inhibitor. In the era of proximity-ligation screens such as APEX2 and BioID, reverting to such a traditional method is quite unusual. The amount of the work that went into isolating the BIF is commendable.
- The identification of acetylated actin as a required component of the CAP inhibitory complex is surprising. This paper is the first to show a role for lysine-acetylated actin and it will be interesting to see what other cellular processes lysine-acetylated actin influences.
Questions for the Authors:
- How did the lab begin working on INF2?
- Why did you choose a classic biochemistry approach to identify the INF2 inhibitor?
- Do you hypothesize that other mechanisms exist for relieving INF2 facilitated-autoinhibition besides the deacetylation of actin?
- Interestingly, both INF2 and CAP can contribute to actin polymerization and depolymerization (through severing). Is this purely coincidental or do you think there is a connection there? On a related note, while you’ve shown that CAP-Actin inhibits INF-2 mediated actin polymerization (both nucleation and elongation), does it also inhibit filament severing?
- Do you think acetylated actin may have broader roles than INF2 regulation?
Posted on: 25 January 2019
doi: https://doi.org/10.1242/prelights.7849
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biochemistry category:
Structural basis of respiratory complexes adaptation to cold temperatures
Lens Placode Modulates Extracellular Matrix Formation During Early Eye Development
Generalized Biomolecular Modeling and Design with RoseTTAFold All-Atom
Also in the cell biology category:
Fetal brain response to maternal inflammation requires microglia
Alteration of long and short-term hematopoietic stem cell ratio causes myeloid-biased hematopoiesis
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
preLists in the biochemistry category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
Preprint Peer Review – Biochemistry Course at UFRJ, Brazil
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biochemistry deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the cell biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |











 (No Ratings Yet)
(No Ratings Yet)