A DNA-based voltmeter for organelles
Preprint posted on 16 January 2019 https://www.biorxiv.org/content/10.1101/523019v1
Article now published in Nature Nanotechnology at http://dx.doi.org/10.1038/s41565-020-00784-1
Electrically excitable organelles? Measuring subcellular membrane potentials with a DNA-based voltmeter
Selected by Robert MahenCategories: biochemistry, bioengineering, biophysics, cell biology, molecular biology, physiology, synthetic biology
Background
Cells across the tree of life use electricity to rapidly transmit signals. Ionic gradients generated across lipid membranes are used to maintain the beating of our hearts, to synchronise the fusion of sperm and egg during fertilization, and in the neural functions that produce our consciousness. Although these voltages are classically measured at the plasma membrane using electrodes, eukaryotic cells also contain internal membrane-bound organelles. Saminathan et al. have built a sensor capable of measuring membrane potentials in intracellular organelles inside living cells using DNA nanotechnology and imaging.
Key findings
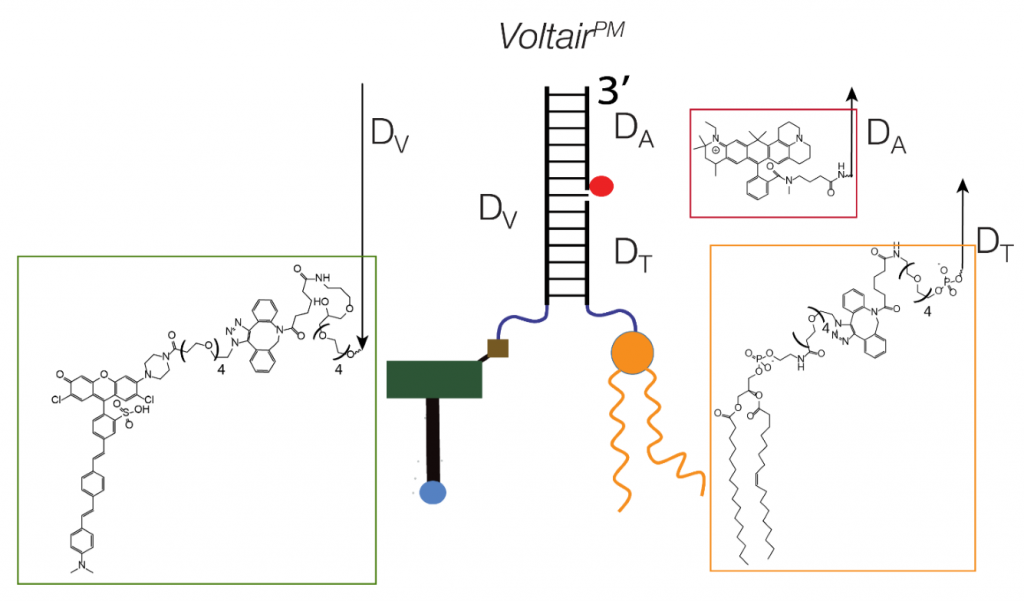
A DNA based device to measure membrane voltages in living cells.
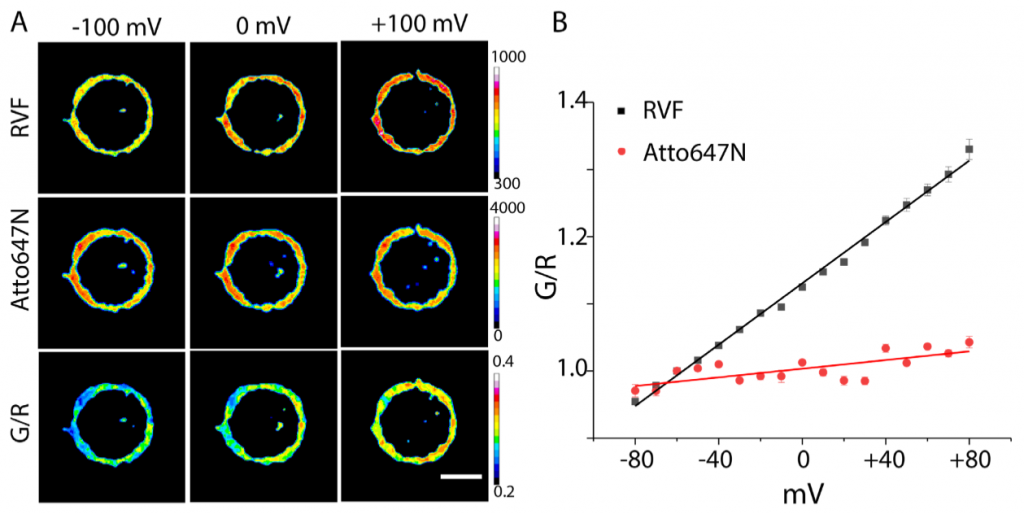
Saminathan et al. term their DNA nanomachine “Voltair” (Fig. 1). Voltair contains two different fluorescent dyes linked at 1:1 stoichiometry using a 38-base pair DNA duplex. The first dye, RVF, changes its fluorescence intensity as a function of potential difference changes along its axis (1, 2), and therefore acts as a voltage sensor. The second dye – Atto647N – is not voltage sensitive, providing a reference for normalisation of fluorescence intensity changes which are not related to voltage. As proof of principle, when whole cell membrane potential is clamped over a range of voltages, Voltair responds with linear changes in fluorescence intensity (Fig. 2). These ratiometric fluorescence intensity values were calibrated against membrane potentials, providing a device to measure absolute voltage in living cells.
Measuring intracellular membrane potentials in diverse cellular locations.
Voltair has a modular structure (Fig. 1), allowing the attachment of different moieties able to target it to distinct cellular membranes. Coupling of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine lipid (POPE) to the probe inserts it in the plasma membrane. It can be trafficked through the endocytic pathway, or alternatively, targeted to recycling endosomes by an aptamer that binds the human transferrin receptor. Localisation to the trans-Golgi network was achieved by expression of furin fused to a single-chain variable fragment recombinant antibody which selectively binds a d(AT)4 sequence in the DNA duplex of the sensor.
Thus, the attachment of different moieties which differentially interact with cellular structures allowed Voltair to report on membrane potentials in different subcellular locations. This showed that membrane potentials differ in different membrane-bound organelles, changing as a function of endosomal maturation in the endocytic pathway, or as a function of mTor localisation at the lysosome.
Why I liked this preprint
Eukaryotic cells are partitioned into membrane-bound organelles with distinct proteomic, chemical and functional composition. Voltair complements existing electrophysiological techniques (3, 4) for measuring membrane potential. As an imaging-based tool, it has the potential to be relatively non-invasive, allows the simultaneous imaging of cellular morphology, and is applicable to high-throughput measurements in populations of cells. Saminathan et al., 2019 show that different membrane-bound organelles have distinct electrochemical compositions. This preprint provides a tool with which to investigate further how membrane bound organelles exploit bioelectricity to regulate their function.
Questions
What are the primary advantages of constructing the device around DNA? Could a similar sensor be realised with a different chemistry?
Cells have elaborate mechanisms to recognise and respond to foreign or damaged DNA. Is it possible that these mechanisms are engaged when Voltair is introduced into a cell?
Are there limitations which currently hamper the probe from being a generic tool able to bind any given cellular target?
References
- Miller EW, Lin JY, Frady EP, Steinbach PA, Kristan WB Jr, Tsien RY. Optically monitoring voltage in neurons by photo-induced electron transfer through molecular wires. Proc Natl Acad Sci U S A. 2012 Feb 7;109(6):2114-9.
- Kulkarni RU, Kramer DJ, Pourmandi N, Karbasi K, Bateup HS, Miller EW. Voltage-sensitive rhodol with enhanced two-photon brightness. Proc Natl Acad Sci U S A. 2017 Mar 14;114(11):2813-2818.
- Hodgkin AL, Huxley AF. Action potentials recorded from inside a nerve fibre. Nature. 1939;144:710–711.
- Lazzari-Dean JR, Gest AMM, Miller EW. Optical determination of absolute membrane potential. 2019. bioRxiv doi 10.1101/519736.
Posted on: 15 February 2019
doi: https://doi.org/10.1242/prelights.8538
Read preprintHave your say
Sign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biochemistry category:
Structural basis of respiratory complexes adaptation to cold temperatures
Lens Placode Modulates Extracellular Matrix Formation During Early Eye Development
Generalized Biomolecular Modeling and Design with RoseTTAFold All-Atom
Also in the bioengineering category:
Scalable and efficient generation of mouse primordial germ cell-like cells
Multi-pass, single-molecule nanopore reading of long protein strands with single-amino acid sensitivity
Discovery and Validation of Context-Dependent Synthetic Mammalian Promoters
Also in the biophysics category:
Structural basis of respiratory complexes adaptation to cold temperatures
Actin polymerization drives lumen formation in a human epiblast model
Learning a conserved mechanism for early neuroectoderm morphogenesis
Also in the cell biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Structural basis of respiratory complexes adaptation to cold temperatures
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
Also in the molecular biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Nanos2+ cells give rise to germline and somatic lineages in the sea anemone Nematostella vectensis
Plant plasmodesmata bridges form through ER-driven incomplete cytokinesis
AND
Plasmodesmata act as unconventional membrane contact sites regulating inter-cellular molecular exchange in plants
Also in the physiology category:
Unlocking the secrets of kangaroo locomotor energetics: Postural adaptations underpin increased tendon stress in hopping kangaroos
Changes in surface temperatures reveal the thermal challenge associated with catastrophic moult in captive Gentoo penguins
Torpor energetics are related to the interaction between body mass and climate in bats of the family Vespertilionidae
Also in the synthetic biology category:
Discovery and Validation of Context-Dependent Synthetic Mammalian Promoters
Genetically encoded multimeric tags for intracellular protein localisation in cryo-EM
Dissecting aneuploidy phenotypes by constructing Sc2.0 chromosome VII and SCRaMbLEing synthetic disomic yeast
preLists in the biochemistry category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
Preprint Peer Review – Biochemistry Course at UFRJ, Brazil
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biochemistry deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the bioengineering category:
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Advances in microscopy
This preList highlights exciting unpublished preprint articles describing advances in microscopy with a focus on light-sheet microscopy.
| List by | Stephan Daetwyler |
Also in the biophysics category:
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
66th Biophysical Society Annual Meeting, 2022
Preprints presented at the 66th BPS Annual Meeting, Feb 19 - 23, 2022 (The below list is not exhaustive and the preprints are listed in no particular order.)
| List by | Soni Mohapatra |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Biophysical Society Meeting 2020
Some preprints presented at the Biophysical Society Meeting 2020 in San Diego, USA.
| List by | Tessa Sinnige |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Biomolecular NMR
Preprints related to the application and development of biomolecular NMR spectroscopy
| List by | Reid Alderson |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
Also in the cell biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |
Also in the molecular biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the physiology category:
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
Also in the synthetic biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Advances in Drug Delivery
Advances in formulation technology or targeted delivery methods that describe or develop the distribution of small molecules or large macromolecules to specific parts of the body.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)
5 years
Sebastian Brauchi
This is actually a nice work from Krishnan lab. But, there’s always more than one way to catch a rabbit…
😉
https://www.biorxiv.org/content/10.1101/578765v1