A Scube2-Shh feedback loop links morphogen release to morphogen signaling to enable scale invariant patterning of the ventral neural tube
Preprint posted on 13 November 2018 https://www.biorxiv.org/content/early/2018/11/13/469239
Size matters: a size-dependent factor that scales sonic hedgehog morphogen gradient in early DV patterning.
Selected by Teresa RayonCategories: developmental biology
Summary:
How do morphogen gradients adapt to size differences? Within populations, individuals vary in size, yet their proportions are similar. This process is known as scale-invariant patterning, and it is a crucial process in development [1,2]. A well-characterized example of this mechanism is the patterning of the neural tube, where it has been shown that embryonic proportions are constant in mouse embryos of different sizes, as well as in embryos of different species. In this preprint, Collins et al. investigate the mechanism by which sonic hedgehog (Shh) morphogen gradient scales in the ventral neural tube in zebrafish embryos of different sizes.
To generate embryos of varying sizes, they surgically remove some cells in the blastula stage (prior to neuroectoderm formation); this rendered embryos of smaller sizes but with constant proportions. Quantifications of a Shh target demonstrate that the response is scaled following embryo reduction.
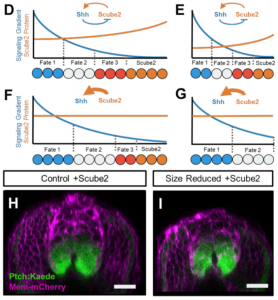
The authors focus their attention on Scube2, a lipid-binding protein required for Shh release non-cell-autonomously, which is expressed in the dorsal and intermediate neural tube. They demonstrate that Scube2 is a diffusible factor distributed throughout the embryo that is secreted from dorsal cells, and it is repressed by Shh signaling.
The authors next wanted to test whether Scube2 expression would scale like Shh reporter genes in sized-reduced embryos. When they measure Scube2 expression in size-reduced embryos, they find that the reduction in Scube2 levels is not scaled: the levels are 50% reduced at DV positions of maximal expression compared to controls, a pattern not found for al scaling invariant genes formerly tested. In addition, when Scube2 is overexpressed in size-reduced embryos, Shh target response is of the same amplitude in embryos of all sizes. This implicates that control of scube2 is responsible for adjusting the Shh signalling gradient in a decreased tissue.
Why I chose the paper:
Embryo development is strikingly robust, and it can cope with variations in size or morphological alterations [3], however the mechanisms that allow embryos to adapt are poorly understood. Collins et al. identify a size-dependent factor that allows the scaling of Shh morphogen gradient in embryos of reduced sizes, adjusting proportionally to the embryonic axis.
The first thing I liked about this work is the surgical method developed in The Megason group to change embryo size and look at size-dependent scaling. They generate perfectly viable and scaled embryos by removing a big percentage of cells at the blastula stage without the need of genetically altering the embryos, which then allows them to perturb different genes.
Their molecular characterization with fish mutants and transgenic reporters is reminiscent of an expander-repressor model [1]. In this model, the morphogen inhibits the expression of an “expander” molecule (Scube2 ), which functions to increase the gradient, holding back morphogen levels at a specific position. What I like about this model is that it does not rely on morphogen diffusion or degradation – that are common molecular properties of proteins regardless of the embryo size – but it relies on the feedback between “expander” and “inhibitor” to continuously adjust the gradient globally. It will be good to see how an expander-repressor model adjusts to their findings and what new predictions we can infer from the model.
Further reading:
Almuedo-Castillo, M., Bläßle, A., Mörsdorf, D., Marcon, L., Soh, G. H., Rogers, K. W., … Müller, P. (2018). Scale-invariant patterning by size-dependent inhibition of Nodal signalling. Nature Cell Biology, 20(9), 1032–1042. http://doi.org/10.1038/s41556-018-0155-7
Ben-Zvi, D., & Barkai, N. (2010). Scaling of morphogen gradients by an expansion-repression integral feedback control. Proceedings of the National Academy of Sciences of the United States of America, 107(15), 6924–9. http://doi.org/10.1073/pnas.0912734107
Umulis, D. M., & Othmer, H. G. (2013). Mechanisms of scaling in pattern formation. Development (Cambridge, England), 140(24), 4830–43. http://doi.org/10.1242/dev.10051
Questions to the authors:
- The neural tube is highly similar in fish and mammals even though there are some differences in the formation of the neural tube. Do the authors think that the expander-repressor model through Scube2 will operate similarly in mammals?
- The size reduction of the embryos occurs early in development, before any Shh has been secreted. Do the authors think their identified mechanism can operate after injury, once the morphogen gradient is ongoing?
- Invariant scaling achieves the same proportions of embryos that vary in size. Do the authors know if the scaling mechanisms have the same tempo in embryos of different sizes?
References:
- Shilo, B.-Z., & Barkai, N. (2017). Developmental Cell Perspective Buffering Global Variability of Morphogen Gradients. Developmental Cell, 40, 429–438. http://doi.org/10.1016/j.devcel.2016.12.012.
- Garric, L., & Bakkers, J. (2018). Shaping up with morphogen gradients. Nature Cell Biology, 20(9), 998–999. http://doi.org/10.1038/s41556-018-0168-2.
- Lawrence, P. A., & Levine, M. (2006). Mosaic and regulative development: two faces of one coin. Current Biology, 16(7), R236–R239. http://doi.org/10.1016/j.cub.2006.03.016.
Posted on: 3 December 2018
doi: https://doi.org/10.1242/prelights.5954
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the developmental biology category:
Temporal constraints on enhancer usage shape the regulation of limb gene transcription
OGT prevents DNA demethylation and suppresses the expression of transposable elements in heterochromatin by restraining TET activity genome-wide
Actin polymerization drives lumen formation in a human epiblast model
preLists in the developmental biology category:
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)