Bacteriophage cooperation suppresses CRISPR-Cas3 and Cas9 immunity
Preprint posted on 10 March 2018 https://www.biorxiv.org/content/early/2018/03/10/279141
Article now published in Cell at http://dx.doi.org/10.1016/j.cell.2018.06.013
Anti-CRISPR phages cooperate to overcome CRISPR-Cas immunity
Preprint posted on 8 March 2018 https://www.biorxiv.org/content/early/2018/03/08/279026
Article now published in Cell at http://dx.doi.org/10.1016/j.cell.2018.05.058
Phages team up to defeat CRISPR immunity - Two new preprints investigate the role of anti-CRISPR proteins during phage infections.
Selected by Fillip PortCategories: epidemiology, evolutionary biology, immunology, microbiology
Background
While CRISPR-based genome engineering dominates the headlines these days, the natural function of CRISPR as a prokaryotic immune system is no less exciting. CRISPR is used by many bacteria and archea to fight off viral infections. It is a pretty effective tool and so phages had to evolve ways to suppress CRISPR immunity in order to be successful. Several phages encode proteins, so called anti-CRISPRs (Acr), which can bind to and neutralize CRISPR effector proteins in vitro. However, what role Acr proteins play in establishing phage infections in vivo is unclear. Phages infecting a CRISPR-expressing prokaryote enter a cell loaded with CRISPR effectors, which can destroy viral genomes within minutes. This raises the question whether Acr proteins, which are only expressed upon phage infection, can really act as effective immunosuppressants to protect the phage genome.
Two recent preprints now explore the role of Acr proteins during bacteriophage infections.
The Preprints / Key findings
The first preprint describes work by Mariann Landsberger and colleagues from Edze Westra and Stineke van Houte’s groups at the University of Exeter, while the second manuscript summarizes work by Adair Borges and others from Joseph Bondy-Denomy’s lab at UC San Francisco. Both groups started by analyzing the effect of different Acr proteins on phage infections of bacterial strains with and without CRISPR immunity. As expected, all phages successfully infected bacterial strains lacking a CRISPR immune system, while CRISPR cleared infections of phages lacking Acr genes. Interestingly, CRISPR-expressing bacteria were also protected from Acr-encoding phages added at low density and only succumbed to the infection when the viral load was high.
So why are Acr proteins an effective antidote to CRISPR at high phage density, but unable to protect phages at low density? One possibility would be that at high densities, bacteria are simultaneously infected by multiple phages and the higher load of viral genomes titrates away CRISPR effectors below a critical threshold. Borges et al. were able to exclude that hypothesis by showing that infections of an Acr phage are not helped by addition of a non-replicative phage strain that does not express an Acr proteins. Furthermore, the preprint by Landsberger and colleagues demonstrates that the critical density for successful infections by phages expressing a strong Acr protein occurs at conditions where simultaneous infections are rare. Instead, both groups propose a model where initial infections cause a immunosuppressed state that lasts long after the initial infection has been cleared. Phages that infect such immunosuppressed cells at a later stage can then successfully replicate. In support of this model, Borges et al. were able to show that transient infections of unsuccessful Acr phages can help subsequent infections to be successful. Furthermore, Landsberger et al. show that bacteria that had previously been infected with Arc phages are more susceptible to the transformation by plasmids for which they carry CRISPR spacers. In addition, both studies find that the phage density required for successful infections is inversely correlated with the affinity of the tested Arc protein for its CRISPR target protein.
Together, these data suggest that Acr proteins expressed by unsuccessful phage infections can induce an immunosuppressed state that can be exploited by subsequent phages infecting the same cell (see figure below from Borges et al.).
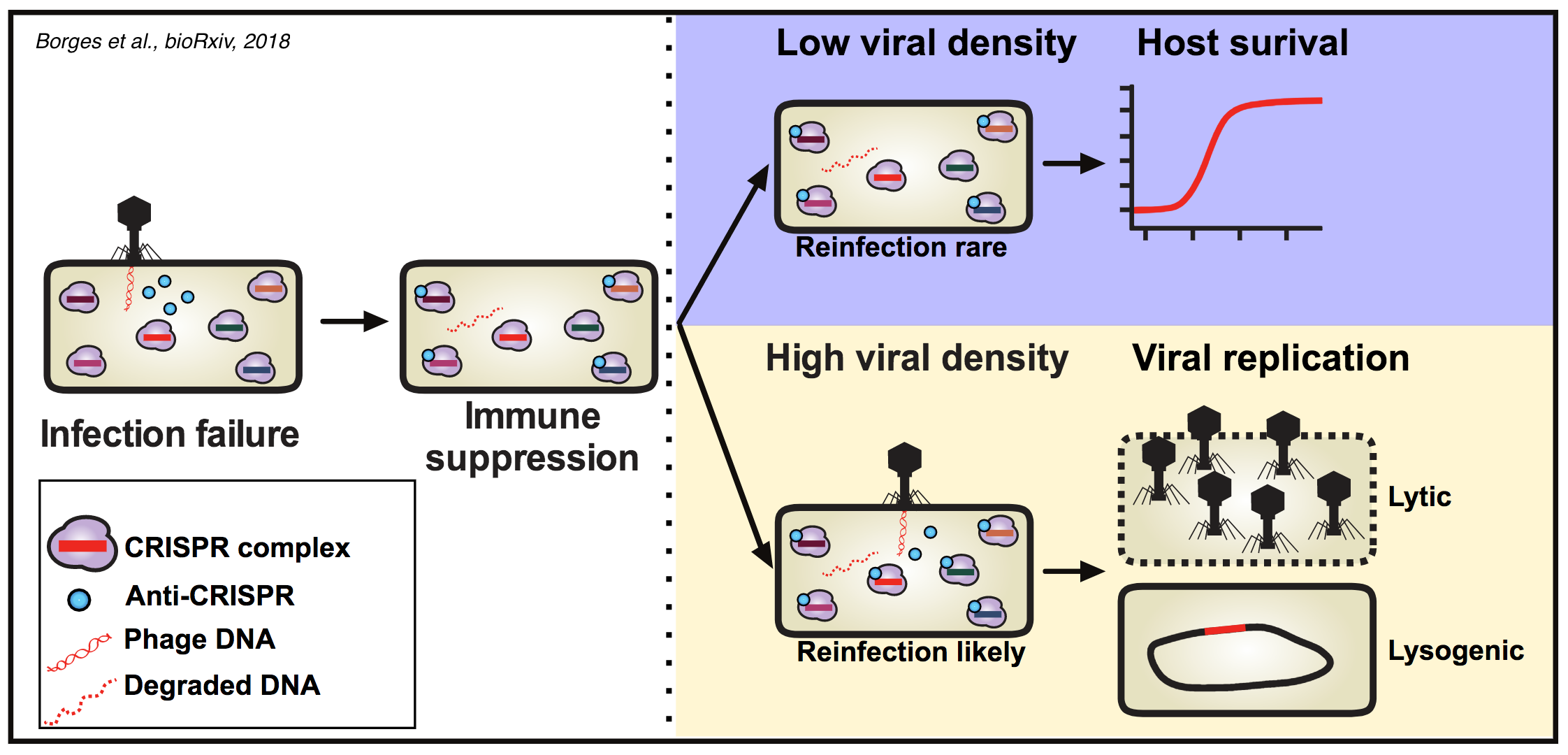
What I like about these pre-prints
The discovery and characterization of anti-CRISPR proteins has been one of the exciting developments in the CRISPR field in the past five years and has lead to a detailed biochemical and structural understanding of some Acr proteins in vitro. These two preprints now widen the focus and investigate the role of Acr proteins during phage infections. After all, as Dobzhansky famously remarked: “Nothing in biology makes sense except in the light of evolution”. With Dobzhansky’s words in mind, what I love about these preprints is that spinning the idea of an evolutionary arms race further readily gives rise to additional hypothesis that can be tested in the future (see below). If lingering Acr proteins cause immunosuppression, what mechanisms might have evolved in some prokaryotes to circumvent that? And if the currently known Acr proteins are unable to prevent elimination of the pioneer phage, what hitherto unknown mechanisms might have evolved in phages that don’t suffer this limitation? It seems certain that the golden age of CRISPR discoveries is far from over.
Besides the science, what I also like about these preprints is the fact that here two groups have investigated the same question and came up with the same answer and have decided to post their finding side-by side on a preprint server. Being afraid of being scooped ranks among the most cited reasons for scientists for being weary of posting preprints. In my view, these two preprints highlight two things: First, truly being scooped is extremely rare. That two teams independently investigating the same question would come up with an identical set of experiments is very unlikely. Rather than taking away from each other, these two manuscripts complement each other and together make a much stronger case for the conclusions than either manuscript alone would have done. Second, posting preprints gives authors control over the timepoint of when their manuscript will be publically available. This can be used to effectively coordinate the simultaneous release of complementary studies from different research teams. Maybe in the future preprint servers could offer additional tools to visibly link manuscripts reporting related findings.
Future directions
These preprints raise a number of questions that might be worth following up in the future.
- Are Acr proteins on their own sufficient to cause lasting immunosuppression in bacteria? For example, would expression of a short pulse of Acr protein expressed from a heterologous plasmid be sufficient to allow subsequent phage infections at low density?
- What is the importance of protein stability for the successful establishment of immunosuppression? Acr proteins that are more stable in the cytoplasm would be expected to provide a longer timeframe for secondary infections to occur. One prediction of that would be that prokaryotes might have evolved mechanisms to directly target Acr proteins for degradation.
- What additional mechanisms might have evolveld in phages to circumvent elimination by CRISPR immune systems? Phages which rely on sequential infections of the same cell might have evolved ways to favor infections of immunosupressed cells. Furthermore, the relative inefficiency of Acr proteins in protecting the phage that encode them makes me wonder whether there might be other yet unidentified Acr proteins that are more potent and might be able to protect a phage that infects a cell with a fully functional CRISPR system. One might imagine enzymatic Acr proteins that can inactivate CRISPR proteins at sub-stoichiometric concentrations.
Posted on: 10 April 2018 , updated on: 11 April 2018
Sign up to customise the site to your preferences and to receive alerts
Register hereAlso in the epidemiology category:
Multimodal interactions in Stomoxys navigation reveals synergy between olfaction and vision
Mapping current and future thermal limits to suitability for malaria transmission by the invasive mosquito Anopheles stephensi
Isolation disrupts social interactions and destabilizes brain development in bumblebees
Also in the evolutionary biology category:
A long non-coding RNA at the cortex locus controls adaptive colouration in butterflies
AND
The ivory lncRNA regulates seasonal color patterns in buckeye butterflies
AND
A micro-RNA drives a 100-million-year adaptive evolution of melanic patterns in butterflies and moths
A complex Plasmodium falciparum cryptotype circulating at low frequency across the African continent
Evolution of tandem repeats in putative CSP to enhance its function: A recent and exclusive event in Plasmodium vivax in India
Also in the immunology category:
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
Spatial transcriptomics elucidates medulla niche supporting germinal center response in myasthenia gravis thymoma
Prenatal inflammation reprograms hyperactive ILC2s that promote allergic lung inflammation and airway dysfunction
Also in the microbiology category:
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
Digital Microbe: A Genome-Informed Data Integration Framework for Collaborative Research on Emerging Model Organisms
Mixed Alkyl/Aryl Phosphonates Identify Metabolic Serine Hydrolases as Antimalarial Targets
preLists in the epidemiology category:
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Also in the evolutionary biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
Also in the immunology category:
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the microbiology category:
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)