Central spindle microtubules are strongly coupled to chromosomes during both anaphase A and anaphase B
Preprint posted on 31 January 2019 https://www.biorxiv.org/content/10.1101/537290v1
Article now published in Molecular Biology of the Cell at http://dx.doi.org/10.1091/mbc.E19-01-0074
The forces from within: the role of central spindle microtubules during chromosome segregation.
Selected by Federico PelischCategories: biophysics, cell biology, molecular biology
Background
Cell division comes in different flavours, namely mitosis and meiosis. In mitosis, two daughter cells arise after duplication and equal partitioning of the genome. Meiosis also involves duplication of the genome but this is followed by two successive rounds of chromosome segregation, generating gametes which are haploid cells (Duro and Marston 2015; Ohkura 2015). Chromosome segregation is achieved by a structure called the spindle, which consists mainly of microtubules (MTs). Spindle MTs contact chromosomes through a multi-protein complex called kinetochore, which assembles on centromeric DNA (Verdaasdonk and Bloom 2011). Centromeres can be localised on specific loci (monocentromeres) or span the whole length of chromosomes (holocentromeres). Regardless of the type of cell division and nature of the centromeres, there is a universal need for chromosome segregation and this always involves MTs. While the most studied mechanism for MT-dependent chromosome movement involves pulling forces generated by MTs emanating from spindle poles making end-on contacts with kinetochores (kMTs), there is evidence for pushing forces that are exerted from within the segregating chromosomes (Khodjakov et al. 2004; Nahaboo et al. 2015; Laband et al. 2017; Vukušić et al. 2017).
Most attention has been given to kMTs, which has provided great insight into the detailed mechanisms of poleward chromosome movement. However, the behaviour of central-spindle MTs, those present within segregating chromosomes, has been far less studied. Early evidence suggested a central-spindle-based mechanism during C. elegans female meiosis (Dumont et al. 2010) and it was later shown that the central-spindle was indeed required for chromosomes segregation (Laband et al. 2017). Very recently, tomographic reconstruction of the worm female meiotic spindle showed that central spindle MTs can make end-on contacts with the inner surface of segregating chromosomes (Redemann et al. 2018). In the present preprint, the authors set out to elucidate the link between central-spindle MT dynamics and chromosome segregation. In order to achieve their goal, they analyse three different spinldes and a variety of methodologies including photoconversion/photobleaching, laser ablation, and electron microscopy.
Key Findings
Yu et al analysed the human mitotic spindle, the C. elegans mitotic spindle, and the C. elegans female meiotic spindle to i) characterise central spindle MT sliding; ii) compare MT sliding and chromosome segregation speeds, and iii) test the impact of ablating central spindle MTs on mitotic and meiotic chromosome segregation (Yu et al. 2019).
One of the key observations is the fact that central spindle microtubules slide apart at the same speed as chromosome segregate (See Figure).
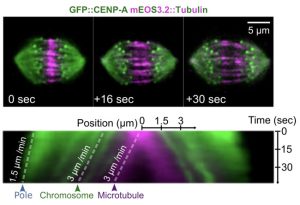
The authors use photoconvertible, mEOS3.2-tagged tubulin to track the sliding behaviour of the central spindle microtubules. In the bottom panel, a kymograph is shown to compare the speeds of MT sliding, chromosome segregation, and spindle poles movement. This kymograph is a very good indication of the key message of the preprint. The figure is reproduced from the preprint’s Figures 2B, C.
By ablating MTs between poles and chromosomes, the authors show that not only chromosomes and central spindle remain somewhat stable, but also that chromosomes segregate. In other words, chromosomes segregate without kinetochore microtubules.
This preprint supports a general a role for central spindle MT dynamics in chromosome segregation. The extent to which this mechanism contributes to chromosome segregation may vary among species. For example, in C. elegans female meiosis where kinetochores are disassembled and seem dispensable for segregation (Dumont et al. 2010; Muscat et al. 2015; McNally et al. 2016; Laband et al. 2017), this central spindle-based mechanism may become the prevailing one.
Overall, the key findings of this preprint are that i) central spindle MTs slide apart at virtually the same speed as chromosomes segregate in the different systems analysed; ii) damaging the central spindle causes an immediate halt in chromosome movement; and iii) spindles contain microtubules with both ends between segregating chromosomes.
What I like about this preprint
I was initially attracted to this preprint due to its close relation to my own research. Technically, I am fascinated by the ablation methodology, especially using femtosecond pulse laser to achieve a complete but reversible halt in chromosome segregation. Also, it seems to be one of those cases in which the authors address a very clear question (not simple to test though) and perform a variety of techniques in different model systems to test it.
Also key to a great piece of work is the fact that this generates new questions which are likely to engage the wide cell division field and will lead to new exciting discoveries.
Open questions
This work opens many questions which will hopefully lead to more exciting research. I will try and highlight some questions that might be of general interest. These questions come to mind after reading this preprint and related articles:
1) What protein(s) regulate central-spindle MT sliding? While the CLASP orthologue CLS-2 plays a role in ‘pushing’ chromosomes away during meiosis (Laband et al. 2017) and mitosis (Nahaboo et al. 2015), how CLS-2 affect microtubule dynamics within the central spindle is not entirely understood.
2) How is MT sliding mechanistically coupled to chromosome movement? In the case of MTs making end-on contacts with the central-spindle facing side of, is there a ‘kinetochore-like’ structure involved?
3) In the case of female meiosis, evidence suggests that CLS-2 plays a role from mid-anaphase (McNally et al. 2016; Laband et al. 2017; Redemann et al. 2018), leaving an open question as to what mechanism(s) are in place to achieve the first step of chromosome separation in the absence of end-on MT-chromosome contacts?
4) It should be noted that in the current preprint as well as in previous work (Nahaboo et al. 2015), some of the conclusions had to be reached after reducing cortical pulling forces (GPR-1/2 depletion). It will be interesting to know to what extent the proposed MT sliding-dependent segregation depends on reduced cortical pulling forces.
References + further reading
Dumont J, Oegema K, Desai A. 2010. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat Cell Biol 12: 894.
Duro E, Marston AL. 2015. From equator to pole: splitting chromosomes in mitosis and meiosis. Genes Dev 29: 109.
Khodjakov A, La Terra S, Chang F. 2004. Laser microsurgery in fission yeast; role of the mitotic spindle midzone in anaphase B. Curr Biol 14: 1330.
Laband K, Le Borgne R, Edwards F, Stefanutti M, Canman JC, Verbavatz JM, Dumont J. 2017. Chromosome segregation occurs by microtubule pushing in oocytes. Nat Commun 8: 1499.
McNally KP, Panzica MT, Kim T, Cortes DB, McNally FJ. 2016. A novel chromosome segregation mechanism during female meiosis. Molecular Biology of the Cell 27:2576.
Muscat, C. C., Torre-Santiago, K. M. , Tran, M. V. , Powers, J. A., and Wignall, S. M. (2015). Kinetochore-independent chromosome segregation driven by lateral microtubule bundles. Elife 4: e06462.
Nahaboo W, Zouak M, Askjaer P, Delattre M. 2015. Chromatids segregate without centrosomes during Caenorhabditis elegans mitosis in a Ran- and CLASP-dependent manner. Molecular Biology of the Cell 26: 2020.
Ohkura H. 2015. Meiosis: An Overview of Key Differences from Mitosis. Cold Spring Harbor Perspectives in Biology 7(5). pii: a015859.
Redemann S, Lantzsch I, Lindow N, Prohaska S, Srayko M, Müller-Reichert T. 2018. A Switch in Microtubule Orientation during C. elegans Meiosis. Current Biology 28: 2991-2997.e2992.
Verdaasdonk JS, Bloom K. 2011. Centromeres: unique chromatin structures that drive chromosome segregation. Nature Reviews Molecular Cell Biology 12: 320.
Vukušić K, Buđa R, Bosilj A, Milas A, Pavin N, Tolić IM. 2017. Microtubule Sliding within the Bridging Fiber Pushes Kinetochore Fibers Apart to Segregate Chromosomes. Developmental Cell 43: 11-23.e16.
Yu C-H, Redemann S, Wu H-Y, Kiewisz R, Yoo TY, Farhadifar R, Muller-Reichert T, Needleman D. 2019. Central spindle microtubules are strongly coupled to chromosomes during both anaphase A and anaphase B. bioRxiv 537290.
Posted on: 12 February 2019 , updated on: 14 February 2019
doi: https://doi.org/10.1242/prelights.8357
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biophysics category:
Structural basis of respiratory complexes adaptation to cold temperatures
Actin polymerization drives lumen formation in a human epiblast model
Learning a conserved mechanism for early neuroectoderm morphogenesis
Also in the cell biology category:
Fetal brain response to maternal inflammation requires microglia
Alteration of long and short-term hematopoietic stem cell ratio causes myeloid-biased hematopoiesis
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Also in the molecular biology category:
Fetal brain response to maternal inflammation requires microglia
Nanos2+ cells give rise to germline and somatic lineages in the sea anemone Nematostella vectensis
Plant plasmodesmata bridges form through ER-driven incomplete cytokinesis
AND
Plasmodesmata act as unconventional membrane contact sites regulating inter-cellular molecular exchange in plants
preLists in the biophysics category:
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
66th Biophysical Society Annual Meeting, 2022
Preprints presented at the 66th BPS Annual Meeting, Feb 19 - 23, 2022 (The below list is not exhaustive and the preprints are listed in no particular order.)
| List by | Soni Mohapatra |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Biophysical Society Meeting 2020
Some preprints presented at the Biophysical Society Meeting 2020 in San Diego, USA.
| List by | Tessa Sinnige |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Biomolecular NMR
Preprints related to the application and development of biomolecular NMR spectroscopy
| List by | Reid Alderson |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
Also in the cell biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |
Also in the molecular biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (No Ratings Yet)
(No Ratings Yet)