Characterization of the Novel Mitochondrial Genome Segregation Factor TAP110 in Trypanosoma brucei
Preprint posted on 26 June 2020 https://www.biorxiv.org/content/10.1101/2020.06.25.171090v1
Article now published in Journal of Cell Science at http://dx.doi.org/10.1242/jcs.254300
Categories: molecular biology
Background:
Trypanosoma brucei, a single-celled Eukaryotic pathogen, causes the serious disease Human African Trypanosomiasis (HAT; Nagana in livestock) in sub-Saharan Africa (1). T. brucei has a single mitochondrion, with mitochondrial genome arranged as series of mini- and maxi- circles of DNA known as a kinetoplast (containing the kDNA). Respiratory chain proteins and rRNAs are encoded on the maxicircles whereas the minicircles encode guide RNAs required for RNA editing of the mitochondrial genes prior to their translation. As the T. brucei mitochondrion is a single copy organelle, it must be duplicated then segregated into the two new daughter cells. kDNA segregation is directed through a complex known as the Tripartite Attachment Complex (TAC), which associates with a structure known as the Basal Body (BB; a microtubule organelle required for assembly of flagella and cilia). The TAC forms de novo during G1-phase beginning from the BB and progressing towards the kDNA following a hierarchical ordering (2,3,4). Though components of the TAC including TAC102 (which was considered the closest protein to the kDNA) have been characterised, much is still unknown about how kDNA segregation is facilitated. Here, the authors uncover a new TAC protein, called TAP110, which interacts with TAC102. Using Ultrastructure Expansion microscopy (U-ExM; 5), they show TAP110 is in fact closer to the kDNA than TAC102 with their cumulated data indicating TAP110 is involved in kDNA segregation.
Key Findings:
- TAP110 localises between the BB and the kDNA.
TAP110 was identified as a putative TAC protein by performing co-immunoprecipitation studies using TAC102 as a bait protein, indicating this protein may play a role in kDNA segregation. By endogenously PTP tagging TAP110 in bloodstream form (BSF; the mammalian infective stage) parasites, the authors show this protein localises across all cell cycle stages to a region between the BB (detected by the antibody YL1/2) and the kDNA (DAPI staining). TAP110 forms two foci prior to kDNA segregation then after segregation, one focus in close proximity to each kDNA network can be seen. Using super resolution microscopy TAP110 and TAC102 signals were shown co-localise. To determine if TAC is needed to localise TAP110 to this region, the authors study TAC formation in cells lacking kinetoplasts (dyskinetoplastic cells). These cells are viable in vitro culture and can be generated by mutating the gamma subunit of the ATPase synthase complex in T. brucei (yL262P) then depleting a TAC protein called p197 by tetracycline (tet) inducible RNAi. After tet removal, the TAC can re-assemble in the absence of kDNA allowing the authors to study this process. Using these cell lines, the authors reveal several findings 1) TAC102 and TAP110 assemble to kDNA region independent of kDNA presence, 2) TAP110 is insoluble in wild-type cells but in dyskinetoplastic cells, TAP110 was detected as part of a putative large 669 kDa complex, 3) by studying TAP110 localisation on isolated flagella, TAP110 was not shown to need the kDNA present to for its localisation to this region but disrupting this interaction does cause a loss of TAC102 and TAP110 to the flagella by over 50 %.
In all, the authors data suggests TAP110 may closely associate with the kDNA.
- TAP110 is closer to the kDNA than TAC102
To investigate the localisation of TAP110 at a higher resolution, the authors used U-ExM in PCF cells. Expansion microscopy (ExM) is a recently established technique which allows the capture of super-resolution images on standard microscopes (to nanoscale structure, 5). Briefly, this technique relies on physically expanding the cells to improve the resolution of factors close to the light diffraction limit. A polymer matrix is introduced to unfixed cells and then denatured. After a first round of expansion into water, cells are immune-labelled. U-ExM is an adapted form of ExM which allows better preservation of ultrastructural components of cells. The authors first validate this strategy in PCF cells by examining tubulin staining. On average, they achieved upwards of 3.5x expansions for the kDNA, the nucleus and the BB. Using U-ExM, they found TAP110 was closer to the kDNA.
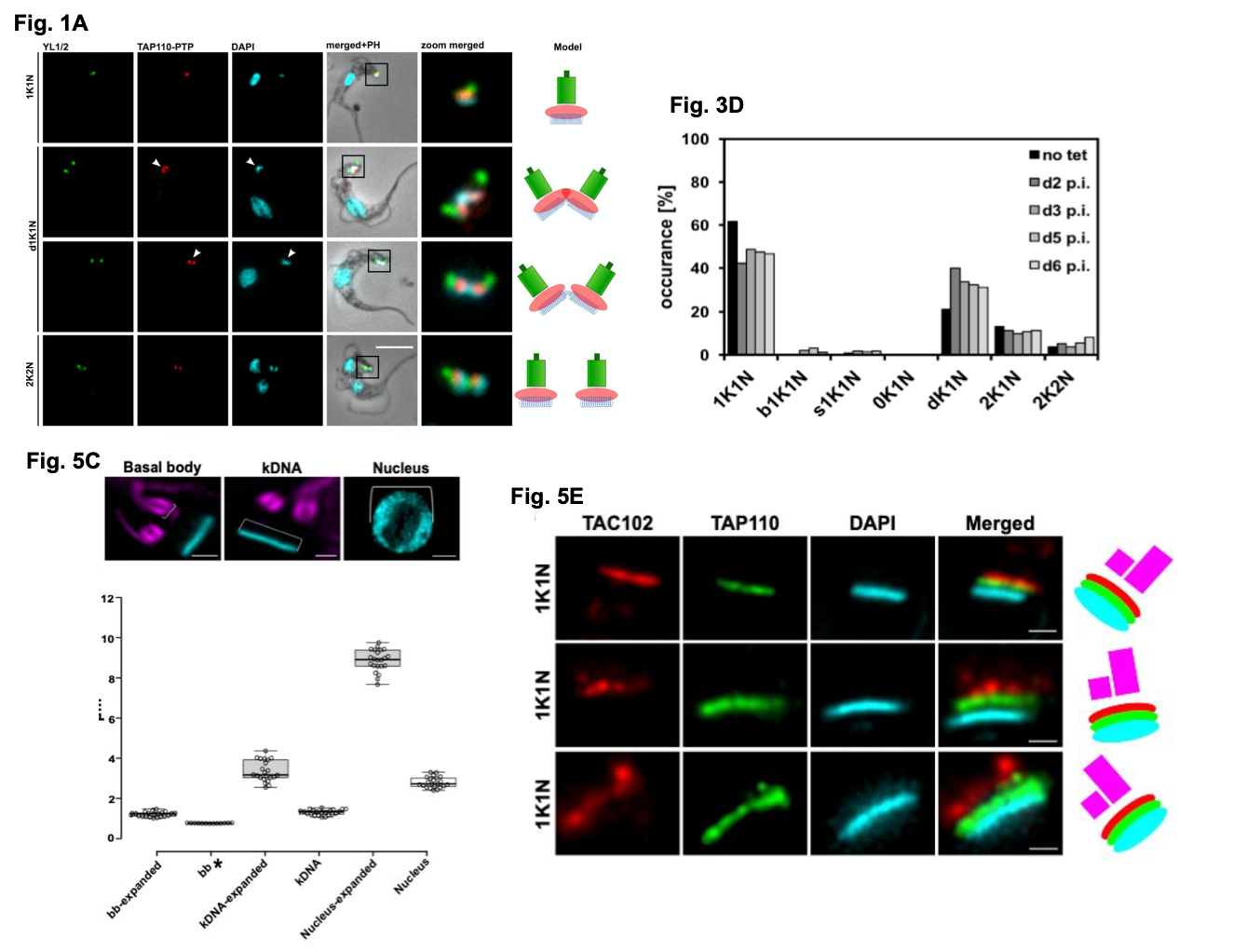
Figure shows a selection of data from Amodeo et al. Fig 1A. details the localisation of TAP110-PTP in representative BSF T. brucei cells at different cell cycle stages. Y1/2 is used to detect the BB with the kDNA and nDNA visualised by DAPI staining. Fig 3D. shows the quantification of kinetoplast (K) and nuclear (N) configurations in the presence or absence of tet inducible TAP110 over expression in PCF cells (d+number = day, p.i = post induction, dK = non-segregated kDNA networks). Fig 5C. (with the images above the graph taken from Fig. 5B) shows the measurements collected from expanded nuclei, BB and kDNA (from U-ExM processing) compared to non-expanded cells. Fig. 5E. shows representative localisations of TAP110, TAC102 and the kDNA in expanded cells using U-ExM. Figures used with author permission.
- Prolonged depletion of TAP110 affects TAC102 in BSF cells
To functionally characterise TAP110 in BSF parasites, the authors depleted TAP110 by tet inducible RNAi then monitored parasite cell cycle and growth. In T. brucei, it is possible to monitor cell cycle progression and kDNA segregation using DAPI staining by examining the ratio of nuclear (N) to kinetoplast (K) DNA. For example, 1N1K cells are G1 cells. It is important to mention that the N and K sub-cycles are uncoupled with the kinetoplast duplicating and segregating prior to nuclear duplication and segregation. Here, the authors found no distinct alterations in growth (vs uninduced cells) and when they compared uninduced cells to cells at 3 days post RNAi induction, no cell cycle disruptions were detected. Exposure to EtBr (ethidium bromide; a DNA intercalating agent which can be used to treat veterinary trypanosomiasis infections) also did not affect growth. However, at 6 days post RNAi induction, the authors noticed increased signal from the kDNA network (~15 % of cells) and either a weak or absent signal for TAC102. When TAC102 was instead depleted by RNAi, the authors found the TAP110 signal was lost in over 80 % of cells indicating TAP110 is closer to the kDNA than TAC102 based on the fact the TAC is assembled hierarchically. If TAC102 was closer than TAP110, TAC102 depletion shouldn’t prevent TAP110 localising.
- Overexpressing TAP110 affects mitochondrial protein abundance
Next, by inducible overexpression of TAP110 fused endogenously to 3x HA tag in the insect stage of the parasite (procyclic form cells; PCFs), the authors show TAP110 has a similar localisation to BSF cells. Only a moderate growth defect was observed upon TAP110 overexpression ~6 day onwards. However, at day 2 post induction, ~ 50 % of cells had duplicated their kDNA network but not segregated it (dK1N) indicating a putative kDNA segregation defect. They found by overexpressing TAP110 in PCF cells and exposing them to heat stress, the growth defect was more pronounced and more abnormal kDNA networks were seen (at 8 days post induction under these conditions).
Using mass spectrometry (MS) to detect changes in the proteome following TAP110 overexpression, the authors uncover (at 2 days post induction), 8 proteins were reduced (4 of which were hypothetical proteins and the other 4 likely mitochondrial) and 4 were enriched (p-value < 0.05, -1 <log2>1). A further two proteins were also included as they met at least one threshold criteria. In all, this data suggest TAP110 overexpression can affect mitochondrial protein levels. One protein of interest was identified using this approach. Up-regulation of a protein known as Tb927.11.6660 was detected. Tb927.11.6660 localises during S-phase of the nuclear cell cycle however, it can localise to the kDNA. Given this protein may interact with TAC102 and possibly TAP110, the authors speculate Tb927.11.6660 could function to link nuclear activities to kDNA activities.
What I liked about this preprint:
What struck me about this preprint was the use of U-ExM. Kinetoplastid parasites are small single-celled organisms and performing detailed, high-resolution microscopy to resolve localisations can be a challenge. The implementation of U-ExM in the field could really improve the access of many laboratories to super-resolution imagining without the dependence upon super-resolution microscopes.
I was also fascinated by the potential connection between the kDNA and the nucleus by the largely uncharacterised gene Tb927.11.6660. What communication occurs between nucleus and the kinetoplast during their respective replication and segregation events is not fully understood. It is exciting to think that follow up study on this newly described protein may help to bridge this gap in our understanding.
Questions for the authors:
Q1: Have you, or do you plan to, investigate the effect of TAP110 overexpression in BSF cells and conversely, the depletion of TAP110 in PCF by RNAi?
Q2: What were the challenges you faced getting U-ExM to work in Trypanosome parasites? You use U-ExM in PCF cells, does U-ExM also work for BSF cells?
Q3: Your data suggests TAP110 is very like a TAC factor but, as mentioned, RNAi or overexpression of TAP110 mildly affects the kDNA suggesting it may not play a major role in kDNA segregation or maintenance. Are you planning on perhaps deleting TAP110, via conventional KO or CRISPR KO, to test TAP110s essentiality?
Q4: Tb927.11.6660 could be an interesting follow up candidate. Looking at trytag.org, the localisation of this protein is linked solely to the nucleolus in PCF cells when C-terminally tagged. Given the localisation in the kDNA and the nucleus observed for BSF cells, do you think this protein could have different functions in different lifecycle stages? This could be interesting to consider due to the lack of a conventional nuclear mitosis-cytokinesis checkpoint in PCF cells, indicating BSF cells may be required to keep tighter control over their duplication and segregation DNA events.
References:
- WHO Trypanosomiasis, human African (sleeping sickness) https://www.who.int/news-room/fact-sheets/detail/trypanosomiasis-human-african-(sleeping-sickness)
- Cavalcanti, D.P., and de Souza, W. The Kinetoplast of Trypanosomatids: From early studies of electron microscopy to recent advances in atomic force microscopy. Scanning (2018).
- Ogbadoyi, E.O., Robinson, D.R., and Gull, K. A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol. Biol. Cell. 14 (2003).
- Hoffman, A. et al. Molecular model of the mitochondrial genome segregation machinery in Trypanosoma brucei. PNAS 115 (2018).
- Gambarotto, D. et al. Imaging cellular ultrastructures using expansion microscopy (U-ExM). Nat. Meth. 16 (2019).
Posted on: 2 July 2020
doi: https://doi.org/10.1242/prelights.22561
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the molecular biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Nanos2+ cells give rise to germline and somatic lineages in the sea anemone Nematostella vectensis
Plant plasmodesmata bridges form through ER-driven incomplete cytokinesis
AND
Plasmodesmata act as unconventional membrane contact sites regulating inter-cellular molecular exchange in plants
preLists in the molecular biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (1 votes)
(1 votes)