Cross-species transcriptomic and epigenomic analysis reveals key regulators of injury response and neuronal regeneration in vertebrate retinas
Preprint posted on 31 July 2019 https://www.biorxiv.org/content/10.1101/717876v2
Article now published in Science at http://dx.doi.org/10.1126/science.abb8598
Fish can fully regenerate cells in the eye after an injury, the chick can do it partially, while mammals lack regenerative potential. The Blackshaw lab explores the pathways activated upon retinal injury in these species.
Selected by Sejal DavlaCategories: neuroscience
Background
In humans and other mammals, retinal injury may lead to blindness as the neural retina tends to form scar tissue rather than regenerate post-injury. In contrast, the zebrafish has a magical ability to regenerate damaged body parts such as eyes, ears and fins, making it a highly desirable vertebrate model to study regeneration.
Muller glia (MG) are specialized glial cells found in the retina of vertebrates where they act as resident stem cells showing a varying degree of neurogenic potential in different species. For example, zebrafish MG pass through a reactive gliosis phase upon tissue-injury, emanating retinal progenitor cells to fully replace the lost neurons whereas MG in post-hatch chicks show only a limited potential for neurogenesis. Mammalian MG, on the contrary, do not show reprogramming ability after tissue injury, but they can be coaxed to reprogram and proliferate using cell-cycle regulators. However, our knowledge about regenerative responses in mammals to date remains insufficient for allowing potential therapeutic interventions.
What gene regulatory programs govern MG proliferation post-injury in fish and chick and how they vary in mammals is the key theme of the preprint from Hoang and Wang et. al. The massive transcriptomic and epigenetic data generated using bulk and single-cell RNAseq and ATAC-seq methodologies survey key regulators of MG-mediated retinal regeneration in zebrafish, chick, and mouse under a variety of injury paradigms and identify novel targets for the improvement of regeneration therapies.
Key findings of the preprint
This preprint provides a comprehensive atlas of genes in MG cells in resting and tissue injury conditions in three species (zebrafish, chicken and mouse) with a varying degree of neurogenic potential in the retina. As expected, these datasets reveal a myriad of genes and markers specific to MG as well as other cell types extant in the retina in each species. Comparison between resting and MG undergoing reactive gliosis within and across species provides an opportunity to identify the key molecular machinery of MG activation in mice.
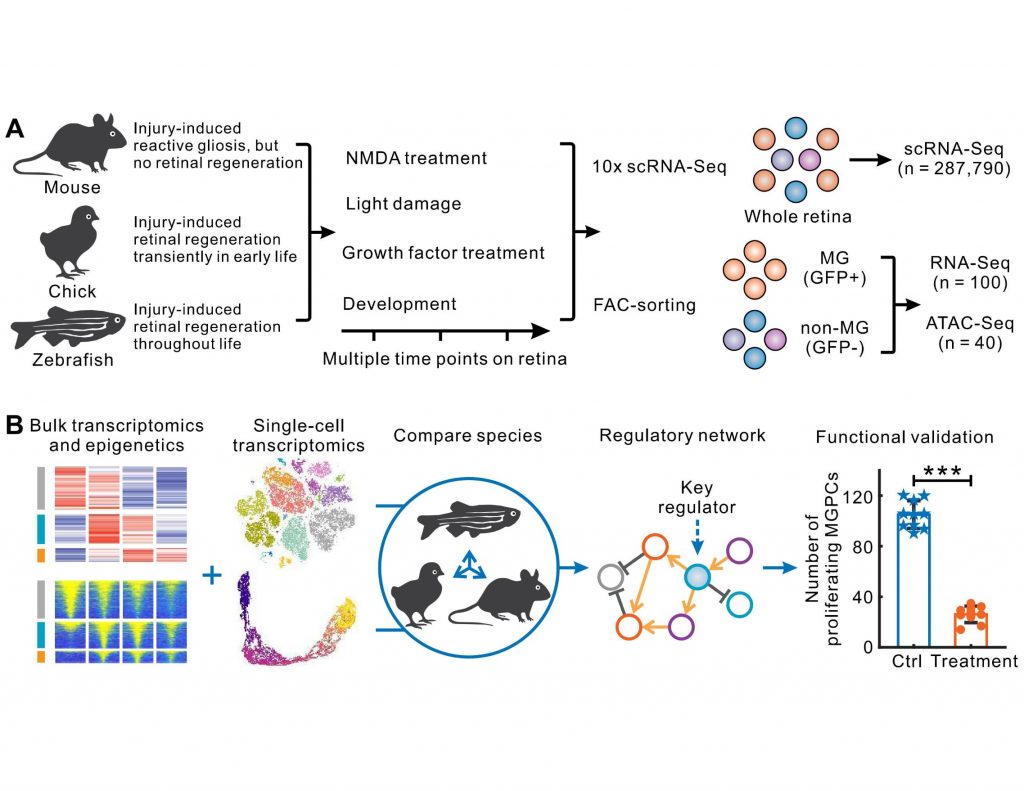
Here, the authors reveal that MG-enriched genes are evolutionarily conserved in all three species. While MG in all three species pass through the reactive gliosis phase after injury, they differ in their injury response trajectories. Zebrafish MG rapidly transition from gliosis to the transcription program that promotes neurogenesis. Detailed gene network analysis shows that quiescence-associated genes in zebrafish MG after an injury activate gliosis genes that, in turn, positively upregulate their expression and suppress quiescence.
Different from zebrafish, post-hatch chick MG show a prolonged gliosis phase but revert to resting-state 48-72 hrs after an injury, where only a small proportion of progenitors generate new neurons. Conversely, mouse MG turn on a unique transcription factor module post-gliosis that suppresses MG proliferation and restores the quiescent state. These gene expression changes further correlate with chromatin configuration state profiled with ATAC-seq where highly expressed genes in both active and resting MG show open chromatin configuration.
Together, these datasets from multiple neuronal injury models exhibit transcriptional similarities suggesting a universal gene regulatory trajectory that defines MG reprogramming in the individual species regardless of the injury protocol used. The changes observed upon neuronal injury remarkably correlate with MG in developing retina and growth condition treatments in zebrafish.
Functional validation of genes identified from transcriptomics in MG reprogramming
This work from the Blackshaw lab is ambitious in its scope and application. The authors used transcriptomics and epigenomics approaches to tease apart the gene network properties of MG during regeneration in different species. Gene network analysis has not only identified universal regulators of MG reprogramming such as Rlbp1, but also revealed injury-regulated genes specific to the species.
While the number of labs using RNA-seq has burgeoned over the last 5 years, very few studies provide the physiological and functional relevance of genes identified from transcriptomics studies. The authors validated the functional significance of novel transcription factors predicted from their transcriptomics data. For example, downregulation of reactive MG genes such as hmga1 and yap1 in zebrafish and FABP genes in chick led to a reduction in the number of new neurons whereas knockdown of transcription factors such as NFI family genes that restore MG quiescence post-injury resulted in increased cell proliferation in the mouse retina.
Future directions and questions for the authors.
This work identifies gene regulatory pathways that either activate MG proliferation or maintain quiescence after tissue injury. Manipulation of these parallel pathways in vivo will uncover the function of hitherto undescribed genes and identify new candidate molecules to enhance regenerative abilities in mammals.
- Did the authors observe heterogeneity in terms of activation of proliferation genes where a population of reactive MG do not turn on gliosis genes in zebrafish? Did the authors distinguish genetic pathway for scar formation in the mouse reactive MG population?
- As the authors highlighted, Ascl1 and Lin28a transcription factors are crucial for MG reprogramming in zebrafish, but mammalian MG do not accumulate these transcription factors that confer neurogenic potential. Are these two genes sufficient to confer neurogenic potential? Do MG proliferation in NFI-deficient mouse and FABP inhibition in chick occur via the activation of Ascl1 and Lin28a?
Posted on: 15 September 2019 , updated on: 16 September 2019
doi: https://doi.org/10.1242/prelights.13802
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the neuroscience category:
Transcriptional profiling of human brain cortex identifies novel lncRNA-mediated networks dysregulated in amyotrophic lateral sclerosis
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
Synergistic olfactory processing for social plasticity in desert locusts
preLists in the neuroscience category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |











 (No Ratings Yet)
(No Ratings Yet)