Defining how Pak1 regulates cell polarity and cell division in fission yeast
Preprint posted on 2 August 2019 https://www.biorxiv.org/content/10.1101/722900v1.full
Article now published in Journal of Cell Biology at http://dx.doi.org/10.1083/jcb.201908017
How does a protein kinase regulate events during cell division? This recent preprint sheds light on how ‘Pak1’ directly influences cell division all the way from initiation to separation in S. pombe.
Selected by Leeba Ann ChackoCategories: cell biology, genetics, microbiology
Background:
Cytokinesis is the process by which a parent cell divides into two or more daughter cells. Cytokinesis can occur either through mitosis or meiosis – both of which are fundamental processes for life. The steps involved in cytokinesis are carefully controlled by higher order actomyosin structures at the cell cortex. Together these structures generate forces that enable the constriction of the contractile actomyosin ring (CAR) and ultimately induce division.
S.pombe follows four essential steps during cell division – i) recruitment of proteins involved in cytokinesis to the cell center at cortical spots called, ‘nodes’, ii) coalescence of these proteins at the nodes into an intact ring (CAR) through actomyosin based interactions, iii) maturation of the ring through recruitment of additional proteins to the CAR, iv) constriction of the CAR thereby enabling cell division via actomyosin forces and cell wall deposition.
Several important players are involved in these four steps of cytokinesis and coordinate various events to promote division. Magliozzi et al. show that the Cell division control protein 42 homolog (Cdc42)-activated polarity kinase, Pak1 directly influences components of the CAR, as well as later stages of cell division by preventing the assembly of ribonucleoprotein granules. Magliozzi et al. identified novel Pak1 substrates that regulate polarized growth, cell division and separation thus, uncovering mechanisms through which the conserved kinase, Pak1 regulates various events during the cell cycle.
Key findings:
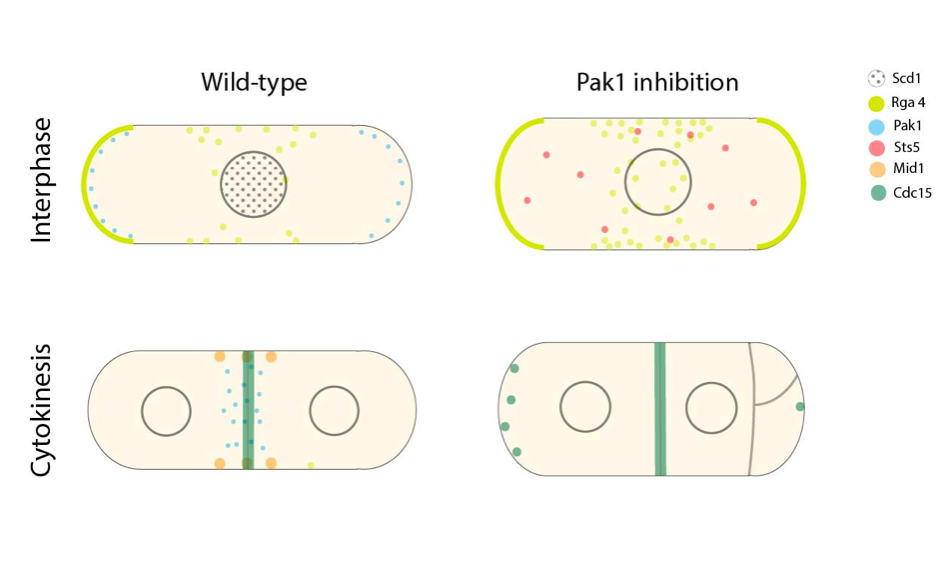
1) During growth, Pak1 oscillates between the two cell ends in a pattern similar to Cdc42 and this is attributed to the localization of Pak1 to the non-constricting rings adjacent to the CAR during septation. At mitotic entry, Pak 1 localizes to the assembling CAR earlier than other cell polarity kinases. Interestingly this localization does not fully overlap with the localization of myosin II regulatory light chain, rlc1 indicating that Pak1 may play a specific role in the coordination of the CAR in space and time.
2) Pak1 mutants take longer to assemble the CAR and this is due to the shorter CAR maturation phase (the period between CAR assembly and constriction). Additionally, Pak1-Mid1/anilin and Pak1-Rng2/IQGAP double mutants exhibit growth deficiencies coupled with cytokinesis defects. Thus, it is likely that due to the localization of Pak1 in the space between nodes and its effect on CAR maturation, the function of Pak1 during early stages of CAR formation is to coalesce nodes into an intact ring.
3) Pak1 influences cell polarization through its ability to directly phosphorylate known cell polarity proteins such as Rga4 and Scd1. In the Pak1 mutant, Rga4 shows increased localization at the cortical puncta and in Pak1-Pom1 (another polarity protein kinase in fission yeast) double mutants, Rga4 is enriched at both cell ends as opposed to a single cell tip, indicating the role of Pak1 in regulating both clustering and tip exclusion of Rga4. Pak1 also regulates the nuclear localization of Scd1. Thus, Pak1 plays a role in maintaining the localization of cell polarity proteins during growth.
4) Pak1 influences cytokinesis through its ability to directly phosphorylate known cytokinesis proteins such as Mid1 and Cdc15. In the Pak1 mutant, the localization of Mid1-Nter is disrupted and consequently the cell exhibits severely mis-localized septa. During cytokinesis, Cdc15 leaves the cell ends and localizes to the CAR. However, in the Pak1 mutant, a fraction of the Cdc15 puncta remain at the cell tips suggesting that Pak1 plays a role in enabling the complete redistribution of Cdc15 to the CAR upon mitotic entry.
5) Complete inhibition of Pak1 leads to cell separation defects. Pak1 influences cell separation through its ability to directly phosphorylate Sts5, which is a protein known to functionally link to septation. Sts5 is normally diffused throughout the cell and upon mitotic entry, it localizes into granules. In the Pak1 mutant, Sts5 localizes as granules throughout the cell cycle. The formation of these granules directly induces septation since the deletion of Sts5 rescues the observed septation defect. Additionally, mutation of the two Pak1 phosphorylation sites in Sts5 lead to granule assembly inside the cells similar to what is observed in the absence of Pak1.
What I liked about this preprint:
Firstly, the preprint was very well written and easy to follow. Secondly, as someone who likes studying the regulation of proteins in-vivo, I was quite excited to see how Pak1 induces phenotypic changes through the spatial and temporal regulation of its substrates. Lastly, I was quite fascinated by the direct link between Pak1 and the production of ribonucleoprotein granules and how this ultimately induces septation defects.
Questions for the authors:
1) There seems to be a correlation between the strength of the loss of function of Pak1 and the aspect ratio of the cells (Fig. S2A and S2B). What is the mechanism behind this observed phenotype?
2) You have shown that the Cdc42 activated, Pak-1 is recruited to the CAR around the same time Cdc42 localizes at the CAR (Fig. 1C). You have also shown that Pak-1 affects the localization of downstream effectors of Cdc42 such as Scd1 (Fig. 6B). Recent work from Hercyk et al. showed that the presence of Scd1 inhibits Gef1 at the division site post ring constriction to enable the timely separation of the daughter cells and Scd1 also prevents the abnormal activation of Cdc42 at the cell sides during cell growth. Could it be possible that the effect of Pak1 on Scd1 localization would in turn affect the localization/function of Cdc42, thereby creating a feedback loop that leads to the defective phenotype observed in Pak1 mutants?
3) You showed that the localization and function of the Mid1-Nter was disrupted in the Pak1 mutant. However, Mid1-mNG was unaffected by the inhibition of Pak1. Is this because the C-terminus region of Mid1 rescues the loss of function of the Mid1-Nter in the Pak1 mutant?
References:
- Magliozzi, J. O., Sears, J., Brady, M., Opalko, H. E., Kettenbach, A. N., & Moseley, J. B. (2019). Defining how Pak1 regulates cell polarity and cell division in fission yeast. bioRxiv. http://doi.org/10.1101/722900
- Loo, T.-H., & Balasubramanian, M. (2008). <em>Schizosaccharomyces pombe</em> Pak-related protein, Pak1p/Orb2p, phosphorylates myosin regulatory light chain to inhibit cytokinesis. The Journal of Cell Biology, 183(5), 785.
- Hercyk, B. S., Rich-Robinson, J. T., Mitoubsi, A. S., Harrell, M. A., & Das, M. E. (2019). A novel interplay between GEFs orchestrates Cdc42 activity during cell polarity and cytokinesis. bioRxiv, 9(2), 364786. http://doi.org/10.1101/364786
Posted on: 10 September 2019
doi: https://doi.org/10.1242/prelights.13692
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cell biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Structural basis of respiratory complexes adaptation to cold temperatures
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
Also in the genetics category:
A long non-coding RNA at the cortex locus controls adaptive colouration in butterflies
AND
The ivory lncRNA regulates seasonal color patterns in buckeye butterflies
AND
A micro-RNA drives a 100-million-year adaptive evolution of melanic patterns in butterflies and moths
A revised single-cell transcriptomic atlas of Xenopus embryo reveals new differentiation dynamics
The genetic basis of novel trait gain in walking fish
Also in the microbiology category:
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
Digital Microbe: A Genome-Informed Data Integration Framework for Collaborative Research on Emerging Model Organisms
Mixed Alkyl/Aryl Phosphonates Identify Metabolic Serine Hydrolases as Antimalarial Targets
preLists in the cell biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |
Also in the genetics category:
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the microbiology category:
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)