Dynamic control of proinflammatory cytokines Il-1β and Tnf-α by macrophages is necessary for functional spinal cord regeneration in zebrafish
Preprint posted on 28 May 2018 https://www.biorxiv.org/content/early/2018/05/28/332197.1
Article now published in Nature Communications at http://dx.doi.org/10.1038/s41467-018-07036-w
To regenerate, or not to regenerate? Researchers shed light on key immune mechanisms and spatiotemporal dynamics that orchestrate spinal cord regeneration in zebrafish.
Selected by Shikha NayarCategories: developmental biology, genetics, immunology
Context and Background
The zebrafish is a well-recognized model system used for vertebrate regeneration research, including, but not limited to, heart, liver, brain, fin, and spinal cord regeneration1. The observation that a robust immune response is elicited upon spinal cord injury is not a novel one2; yet key mechanisms, cell types, and spatiotemporal dynamics are still to be elucidated in a vertebrate system. Macrophages, neutrophils, and microglia are fundamental innate immune cell types that elicit an immediate immune response upon injury, yet their dynamics are rapid and thus difficult to study. The zebrafish embryo is a unique model system as it only possesses a functional innate, but not adaptive, immune system at embryonic and larval stages. This makes it highly effective for dissecting mechanisms of innate immune cell infiltration and axon growth during spinal cord regeneration in vivo (Figure 1).
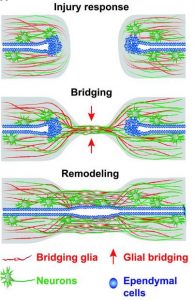
Figure 1. Schematic of multistep process of spinal cord regeneration in zebrafish (Figure 1a from Mokalled et al., 2016).
Key findings
How do infiltrating immune cells control cytokine dynamics to promote mechanisms of regeneration? In this preprint, Tsarouchas et al use the zebrafish embryo to show that peripheral macrophages control a delicate balance of pro-regenerative and pro-inflammatory cytokine production to promote successful axon regeneration. They show that macrophages, but not neutrophils, are necessary for the regulation and maintenance of this cytokine balance. They conclude that an induction of tnfα but reduction of il1β is necessary and sufficient to sustain successful axon regeneration through development. Importantly, the tight spatiotemporal control of cytokine induction was key for long-term regenerative success; early induction of a pro-inflammatory response, but a switch to an anti-inflammatory response after 24 hours is needed for effective regeneration. Notably, macrophage mutants were unable to switch between these biphasic cytokine states, resulting in unsuccessful axon regeneration.
Importance of this paper
The textbook view in immunology is that sustained induction of pro-inflammatory cytokines, such as tnfα, is responsible for chronic injury and a failure to elicit repair responses. This study importantly challenges this dogma by dissecting the dynamics of cytokine production through injury and development, to show that appropriate and timely induction of pro-inflammatory cytokines can in fact play a key role in repair. Not only will these findings inform future directions in regeneration research, but they could shape our view of immune dynamics in other disease models, such as Inflammatory Bowel Disease, that also currently view cytokines like tnfα as detrimental to healing and repair processes.
Open questions
The authors do discuss the intricacies of axon bridging as a potential therapeutic strategy, as shown in neutrophil-depleted mutants, however this idea could be further explored. How does axon bridging naturally take place, and what immune mechanisms are necessary to orchestrate the process? How do patients with defects in innate immune populations such as neutrophils and macrophages, resolve injury induced by noxious stimuli? What are the cytokine levels and spatiotemporal dynamics in patients with dysfunctional macrophages? Could these patients be targeted for “personalized cytokine induction”? Further, this study is carried out with the goal of dissecting primarily innate immune mechanisms of repair; how would these dynamics be altered with the emergence of a functional adaptive immune system in vivo later in development? Investigating the intersection of immunology, regeneration, and the use of novel model systems, will help us answer these questions and provide new avenues for translational approaches in spinal cord regeneration.
References
- Gemberling A, Bailey TJ, Hyde DR, Poss KD. The zebrafish as a model for complex tissue regeneration. Trends in Genetics. 2013;29(11):611-620. doi:10.1016/j.tig.2013.07.003.
- Mokalled MH, Patra C, Dickson AL, Endo T, Stainier DYR, Poss KD. Injury-induced ctgfa directs glial bridging and spinal cord regeneration in zebrafish. Science. 2016;6312(354):630-634. doi:10.1126/science.aaf2679.
- Trivedi A, Olivas AD, Noble-Haeusslein LJ. Inflammation and Spinal Cord Injury: Infiltrating Leukocytes as Determinants of Injury and Repair Processes. Clinical neuroscience research. 2006;6(5):283-292. doi:10.1016/j.cnr.2006.09.007.
Posted on: 30 July 2018
doi: https://doi.org/10.1242/prelights.3930
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the developmental biology category:
Temporal constraints on enhancer usage shape the regulation of limb gene transcription
OGT prevents DNA demethylation and suppresses the expression of transposable elements in heterochromatin by restraining TET activity genome-wide
Actin polymerization drives lumen formation in a human epiblast model
Also in the genetics category:
Temporal constraints on enhancer usage shape the regulation of limb gene transcription
A long non-coding RNA at the cortex locus controls adaptive colouration in butterflies
AND
The ivory lncRNA regulates seasonal color patterns in buckeye butterflies
AND
A micro-RNA drives a 100-million-year adaptive evolution of melanic patterns in butterflies and moths
A revised single-cell transcriptomic atlas of Xenopus embryo reveals new differentiation dynamics
Also in the immunology category:
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
Spatial transcriptomics elucidates medulla niche supporting germinal center response in myasthenia gravis thymoma
Prenatal inflammation reprograms hyperactive ILC2s that promote allergic lung inflammation and airway dysfunction
preLists in the developmental biology category:
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the genetics category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Also in the immunology category:
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)