Dynamics and interactions of ADP/ATP transporter AAC3 in DPC detergent are not functionally relevant
Preprint posted on 10 May 2018 https://www.biorxiv.org/content/early/2018/05/10/317669
Major concerns with the integrity of the mitochondrial ADP/ATP carrier in dodecyl-phosphocholine used for solution NMR studies
Preprint posted on 25 May 2018 https://www.biorxiv.org/content/early/2018/05/25/329490
Article now published in Nature Structural & Molecular Biology at http://dx.doi.org/10.1038/s41594-018-0125-6
Two preprints question the structural details of an ADP/ATP transporter's mechanism and call for more thorough controls to verify membrane protein functionality
Selected by Reid AldersonCategories: biochemistry, biophysics, molecular biology
Context
Membrane proteins (MPs) comprise 15-25% of the human proteome, yet collectively sum to less than 1% of the deposited three-dimensional structures in the Protein DataBank (PDB). Such a paucity of structural information manifests from difficulties that can accompany the biochemical preparation of purified MPs, ranging from complications with faithful abstraction from the native membrane to misfolding after extraction from insoluble inclusion bodies. Moreover, despite the common view of MPs existing in one of two end states (i.e. “open” or “closed” transporters, “active” or “inactive” G-protein coupled receptors), MPs can populate a wide range of conformations that dynamically exchange on various timescales, spanning pico-nanoseconds, micro-milliseconds, or far longer than hours. Such dynamics can often hinder crystallization attempts for structural analysis by X-ray crystallography. Despite the current lack of structural knowledge pertaining to MPs, many FDA-approved drugs target them. Thus, a holistic understanding of membrane protein structures, dynamics, and interactions in native membrane environments, or reliable membrane mimetics, proves critical.
A significant factor in structural studies of MPs is the membrane environment. As pointed out in a recent review1, nearly 80% of MP structures determined to date have used detergents, which form small micelles that present a very different environment to lipids. However, recent work has revealed that many MPs embedded in such micelles, for instance of the commonly used detergent dodecyl phosphocholine (DPC, also known as Foscholine-12), are no longer functional or folded correctly1.
Thus, when choosing a membrane mimetic system, how does one ensure that a reconstituted MP is functionally relevant? The most rigorous demonstration is an assay that directly monitors biochemical activity: for example, the transport of a substrate or other reporter molecule. In the absence of such an assay, researchers typically default to the demonstration of the MP binding to a relevant ligand, generally by measuring a dissociation constant (Kd) and comparing this to values obtained by other groups who have used native membranes. In the preprints discussed below, two reproduction studies are conducted pointing out significant flaws in this practice, and show that the presumed functionality of MPs should be interpreted with caution when using detergents.
The preprints
Two recent preprints conducted reproducibility studies on the structural and functional integrity of an AAC transporter (AAC3) in DPC2,3. AAC3 functions to exchange ADP for ATP across the inner mitochondrial membrane, an essential process for the cell. This membrane protein had previously been crystallized in the presence of a strong inhibitor (CATR) and was shown to adopt a locked conformation, termed the “c-state”4,5. As part of its transport mechanism, AAC3 populates a second conformation known as the “m-state”, yet structural evidence for this state has remained elusive.
In 2015, a paper by Chou and colleagues6 in Nature Structural & Molecular Biology reported using NMR spectroscopy that, in DPC, the yeast version of AAC3 (yAAC3) could be captured in a dynamic equilibrium involving two states: the c-state and a transiently populated conformation, which was inferred to be the m-state (or similar). Critically, their results hinged on yAAC3 remaining correctly folded and fully functional while embedded in the detergent. The integrity and correct folding of the yAAC3 sample was determined by binding of the inhibitor (CATR) and substrate (ADP) as measured by isothermal titration calorimetry (ITC) or NMR6.
Preprints from Schanda and colleagues2 at the Institut de Biologie Structurale in Grenoble (France) and Kunji and coworkers3 at the Mitochondrial Biology Unit in Cambridge (UK) challenged the notion that yAAC3 is correctly folded and functional in DPC. Rather, the authors convincingly demonstrate that yAAC3 is misfolded and not functional in the presence of DPC.
First, both King et al.3 and Kurauskas et al.2 noted that the Kd values for CATR binding to yAAC3 reported by Chou and colleagues were nearly three-to-four orders of magnitude larger than the commonly accepted literature values (5–300 nM). Second, Schanda and colleagues examined the raw data used to determine that the substrate (ADP) binds specifically, and found residues all over yAAC3, including those more than 20 Å distant from the ADP binding site, were impacted by ADP binding2. While this could reflect allosteric structural changes, a recent publication noted that AAC3 embedded in DPC bound identically to ATP and GTP, despite preference for the former in the native membrane5. Such a loss of specificity implies an incorrect tertiary structure. Finally, King et al.3 measured melting temperatures of yAAC3 in DPC and other detergent systems (Figure 1A). In the native mitochondrial membrane, yAAC3 unfolded near 40°C; however, the same experiment performed in DPC revealed a large fraction of unfolded protein already present at ambient temperature, which exhibited no unfolding cooperativity3. Similar results were obtained by NMR3. Taken together, these data suggest that yAAC3 solubilized in DPC is incorrectly folded and binds to CATR and ADP non-specifically2,3. King et al. note that a non-specific interaction could be electrostatic in nature, as yAAC3 has an isoelectric point of 9.82, and therefore contains excess positive charge, whereas ADP and CATR are both negatively charged at neutral pH3.
Having established that ADP and CATR binding in fact reflects non-specific interactions, Schanda and colleagues2 re-examined the previously published NMR data used to measure conformational exchange in yAAC3 between the c-state and putative m-state (Figure 1B). The experiment used for such analysis is called Carr-Purcell-Meiboom-Gill relaxtion dispersion (CPMG RD, reviewed here), and involves the measurement of an NMR parameter (transverse relaxation rate, termed the effective R2) as a function of the number of radiofrequency pulses applied during a fixed delay. In the presence of conformational exchange between two or more states on the micro-to-millisecond timescale, CPMG RD experiments will reveal a dependence of R2 on the number of applied radiofrequency pulses (Figure 1B). Subsequently fitting these data to the appropriate equations yields the rate of conformational exchange, the relative populations, and (qualitative) residue-specific information about structural differences between the two forms. For example, such experiments have been deployed to determine accurate solution structures of otherwise invisible protein folding intermediates that exchange with a folded conformation on the millisecond timescale7.
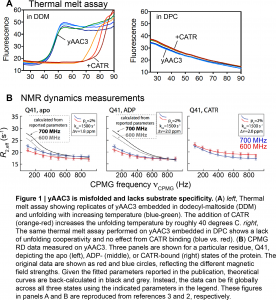
In the original paper, Chou and colleagues6 found via CPMG RD that the rate of conformational exchange varied by roughly 20-fold between the inhibitor- and substrate-bound forms of yAAC3, and reported large structural differences between the two states (for NMR spectroscopists, 15N |delta omega| > 5 ppm). However, Kurauskas et al. re-fit the raw CPMG RD data in free state and the inhibitor- and substrate-bound forms of yAAC3 (Figure 1B), finding that no difference existed in the rate of conformational exchange (1500 s-1 for all forms), with significantly smaller structural differences than initially reported (15N |delta omega| values around 2 ppm). Finally, similar millisecond exchange dynamics that were independent of added substrate/inhibitor were obtained when analyzing conformational exchange in mutants of AAC3 that are incapable of ADP/ATP exchange, indicating that there is no link between dynamics and transport8.
In conclusion, these two new preprints from the Schanda and Kunji groups demonstrate that the yAAC3 membrane protein is not functional and misfolded when embedded in DPC. These studies highlight the difficulty and importance of choosing the right membrane mimetic (when not using native membranes). In addition, these works provide a platform for diagnosing future issues that arise from misfolded MPs (e.g. non-specific ligand binding, lower Kd than expected, pervasive millisecond motions), in particular those analyzed by NMR spectroscopy.
My opinion
These two new preprints highlight the importance of independent replication studies that seek to verify, challenge, or question biological implications obtained from biophysical experiments. While a few examples exist in the published literature, there are certainly not enough of such studies. Encouragingly, a replication preprint was recently highlighted on preLights. The bioRxiv will help to remove the stigma associated with publishing replication studies, and will provide a convenient platform for the publication of replication studies in lieu of publication in traditional journals. Likewise, PLOS Biology has instituted an initiative to publish papers from groups who have recently been “scooped”, which should also increase the number of papers that report data on the same system.
While the two preprints discussed here present negative results, their findings will together advance the field of AAC structural biology, and hopefully will deter future researchers from using DPC as a reconstitution system. More generally, the preprints from the Schanda and Kunji groups highlight the importance of selecting the appropriate membrane mimetic system and then verifying that the embedded MP retains functional and structural integrity.
As a practicing NMR spectroscopist who enjoys dynamical measurements, the experiment that I most enjoyed was actually a data analysis method to obtain insight into micro-to-millisecond motions. Schanda and colleagues noted that the CPMG RD data from Chou and coworkers could be described by a single set of parameters for all three conditions (apo-AAC3, CATR-bound AAC3, ADP-bound AAC3), and showed that these data could indeed be fitted globally. This means that the AAC3 exchange process on the micro-to-millisecond timescale, which is probed in the NMR experiment, is insensitive to added substrate or inhibitor, and is therefore not a transport filter.
References
- Chipot C, et al. Perturbations of native membrane protein structure in alkyl phosphocholine detergents: a critical assessment of NMR and biophysical studies. (2018) Chem. Rev. 118: 3559-607.
- Kurauskas V, Hessel A, Dehez F, Chipot C, Bersch B, Schanda P. Dynamics and interactions of ADP/ATP transporter AAC3 in DPC detergent are not functionally relevant. (2018) bioRxiv
- King MS, Crichton PG, Ruprecht JR, Kunji ERS. Major concerns with the integrity of the mitochondrial ADP/ATP carrier in dodecyl-phosphocholine used for solution NMR studies. (2018) bioRxiv
- Pebay-Peyroula E, Dahout-Gonzalez C, Kahn R, Trezeguet V, Languin GJ, Brandolin G. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. (2003) Nature 426: 39-44.
- Ruprecht JJ, Hellawell AM, Harding M, Crichton PG, McCoy AJ, Kunji ERS. Structures of yeast mitochondrial ADP/ATP carriers support a domain-based alternating-access transport mechanism. (2014) Proc. Natl. Acad. Sci. USA 111: E426-34.
- Bruschweiler S, Yang Q, Run C, Chou JJ. Substrate-modulated ADP/ATP-transporter dynamics revealed by NMR relaxation dispersion. (2015) Nat. Struct. Mol. Biol. 22: 636-41.
- Hansen DF, Vallurupalli P, Kay LE. Using relaxation dispersion NMR spectroscopy to determine structures of invisible, excited protein states. (2008) J. Biomol. NMR. 41: 113-20.
- Kurauskas V, Hessel A, Ma P, Lunetti P, Weinhaupl K, Imbert L, Brutscher B, King MS, Sounier R, Dolce V, Kunji ERS, Capobianco L, Chipot C, Dehez F, Bersch B, Schanda P. How detergent impacts membrane proteins: atomic-level views of mitochondrial carriers in dodecylphosphocholine. (2018) J. Phys. Chem. Lett. 9: 933-8.
Posted on: 23 July 2018 , updated on: 30 July 2018
Sign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biochemistry category:
Structural basis of respiratory complexes adaptation to cold temperatures
Lens Placode Modulates Extracellular Matrix Formation During Early Eye Development
Generalized Biomolecular Modeling and Design with RoseTTAFold All-Atom
Also in the biophysics category:
Structural basis of respiratory complexes adaptation to cold temperatures
Actin polymerization drives lumen formation in a human epiblast model
Learning a conserved mechanism for early neuroectoderm morphogenesis
Also in the molecular biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Nanos2+ cells give rise to germline and somatic lineages in the sea anemone Nematostella vectensis
Plant plasmodesmata bridges form through ER-driven incomplete cytokinesis
AND
Plasmodesmata act as unconventional membrane contact sites regulating inter-cellular molecular exchange in plants
preLists in the biochemistry category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
Preprint Peer Review – Biochemistry Course at UFRJ, Brazil
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biochemistry deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the biophysics category:
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
66th Biophysical Society Annual Meeting, 2022
Preprints presented at the 66th BPS Annual Meeting, Feb 19 - 23, 2022 (The below list is not exhaustive and the preprints are listed in no particular order.)
| List by | Soni Mohapatra |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Biophysical Society Meeting 2020
Some preprints presented at the Biophysical Society Meeting 2020 in San Diego, USA.
| List by | Tessa Sinnige |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Biomolecular NMR
Preprints related to the application and development of biomolecular NMR spectroscopy
| List by | Reid Alderson |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
Also in the molecular biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (No Ratings Yet)
(No Ratings Yet)