Engineering Phage Host-Range and Suppressing Bacterial Resistance Through Phage Tail Fiber Mutagenesis
Preprint posted on 11 July 2019 https://www.biorxiv.org/content/10.1101/699090v1
Article now published in Cell at http://dx.doi.org/10.1016/j.cell.2019.09.015
Phagebodies and the beast of antibiotic resistance: engineering of bacteriophage tails alters host ranges and prevents development of bacterial resistance
Selected by Zhang-He GohCategories: microbiology, synthetic biology
Background of preprint
Research into bacteriophages may be a tale as old as time, but the rise of antibiotic resistance in the past decade has led to a rebound in research into using bacteriophage therapy to mitigate this looming threat [1]. Three advantages of using bacteriophages exist: (a) The effectiveness of bacteriophages are not distinct from mechanisms leading to antibiotic resistance, (b) bacteriophages have already been shown to be effective in the clinic, albeit on a small pool of patients [2,3], and (c) the selectivity of phages for select bacteria reduces their ecological effect—a major cause driving the rise of resistance to broad-spectrum antibiotics in the first place.
However, phages are generally not well characterised, and this problem is compounded in phages developed by natural evolution. Therefore, to overcome these challenges involving phage characterisation, Yehl et al. mutated phage regions—described as the “complementary determining regions (CDR)-like loops at the tip of the tail fibre”—that are necessary for host recognition to modulate host specificity.
Key findings of preprint
(A) Construction of phage libraries with genetic variability in host-range determining regions (HRDR)
Yehl et al. first showed that natural phage evolution is insufficient to suppress resistance by co-culturing the phage T3 with its natural host, E. coli B (preprint Fig. 2C). The authors then generated a large diversity of phages by randomising the DNA coding for the loop sequences in HRDRs. Seven libraries with different randomisations were constructed by randomising the codons in various loops and compared to the wild type phage (preprint Fig. S2B).
The authors then characterised these libraries by quantifying their diversity (preprint Fig S2B):
- Libraries targeting 4 codons saturated the theoretical DNA sequence space,
- While libraries targeting 5 codons did not saturate the theoretical DNA sequence space, they still saturated the protein sequence space after accounting for redundancy in the genetic code, and
- Libraries targeting more than 5 codons did not saturate both the DNA and protein sequence spaces.
During library construction, the authors noted that the limiting step for library diversity is the transformation yield (preprint Fig. S2C).
(B) Loop randomisation and mutations can delay or prevent bacterial resistance
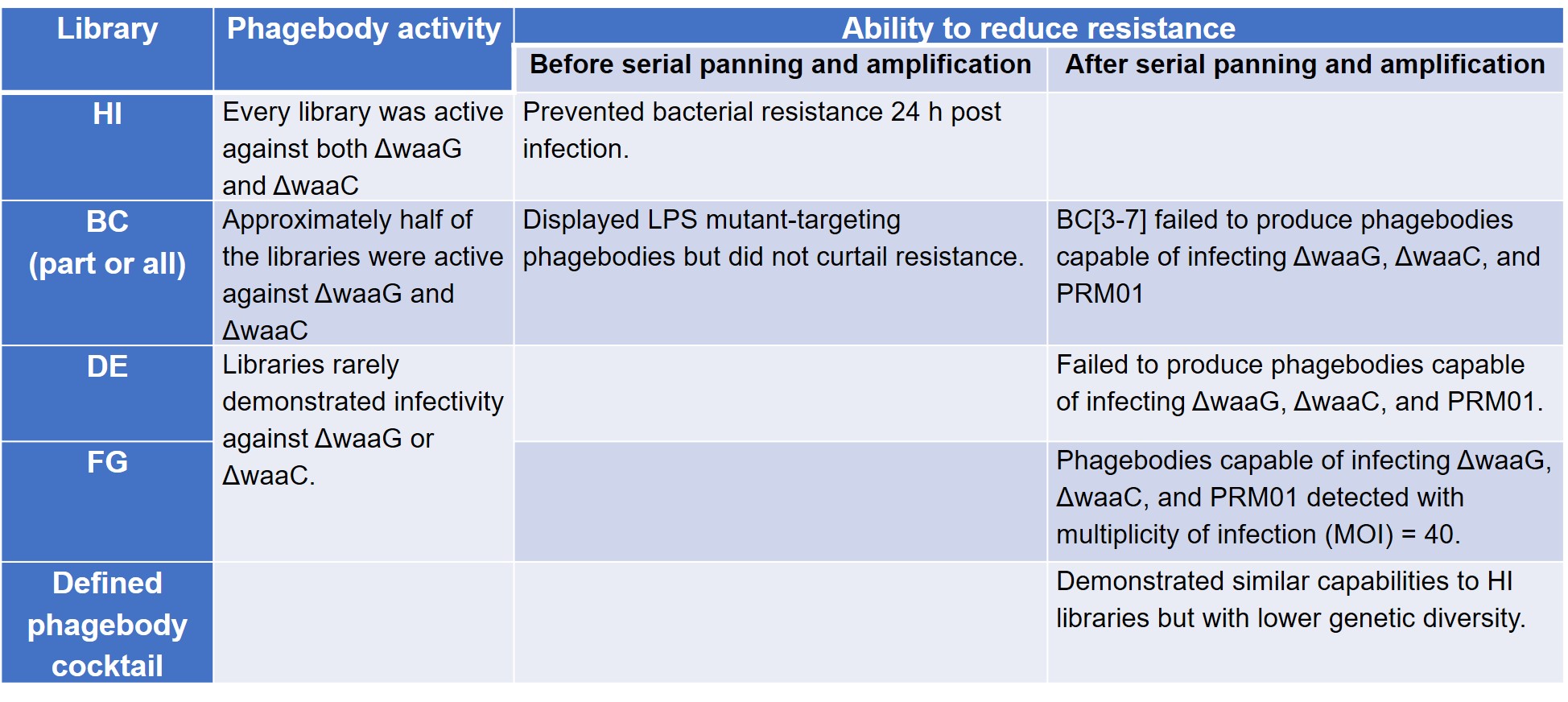
To test their hypothesis that loop randomisation creates functional diversity, Yehl et al. screened these phagebody libraries on T3-resistant ΔwaaG and ΔwaaC LPS mutant strains to prove that they were active. The authors then compared the ability of these phage libraries to reduce the development of resistance to wild-type T3, conducted serial panning and amplification in three mutant host strains—ΔwaaG, ΔwaaC, and Phage Resistant Mutant 01 (PRM01)—to check for rare or poorly growing phagebodies, and tested a defined cocktail comprising mutants enriched from the panning experiment (preprint Table S3).
The results of these four sets of experiments are summarised below (Table 1).
Table 1. Phagebody library activity and mitigation of resistance
(C) Characterisation of selected phagebodies
Yehl et al. isolated phagebodies and compared them to naturally-occurring mutants of T3 (NM), comparing their genomes, host range, and killing efficiency. The authors found that most NMs displayed a broad host-range for the BL21 bacterial panel, while phagebodies exhibited a more diverse range of host-range phenotypes. This observation allowed the authors to explore how physicochemical properties such as charge, hydrophilicity, and hydrophobicity of the respective loop regions correlate with host-range.
Yehl et al. then tested the ability of the phagebodies to suppress the development of resistance, finding only a qualitative link between host-range and killing efficiency, though there was a correlation between the hydrophilicity of the HI loop and resistance suppression. Finally, the authors tested an engineered phenobody, PB10, in a mouse skin infection model, concluding that it could clear ΔwaaG and ΔwaaC from the wounds of these mice.
What I like about this preprint
In their preprint, Yehl et al. demonstrate a method to generate and screen huge phagebody libraries, and use it to identify engineered phages that are able to suppress bacterial resistance to phage infection. Other than proving the potential efficiency of these phages both in vitro and in vivo, the authors also show how a carefully defined cocktail of these phages can aid in safety, thus making such a potential therapy more palatable to regulators.
I felt that this preprint is particularly exciting because it offers man another avenue of development even as we continue our crusade against the rising tide of antimicrobial resistance. It is also forward-looking: the authors describe a method to target potentially phage-resistant mutants even before the field of phage therapy has fully matured. Incorporating these considerations into future designs of phage therapies may offer scientists and healthcare professionals the opportunity to exercise better stewardship of this up-and-coming resource.
The third interesting aspect to this preprint is that this method was inspired by antibody specificity engineering. The explosion of data has ushered in a new era of interdisciplinary science, and this is another example of how one field of protein engineering can be impacted by another. To paraphrase Mrs Potts from Beauty and the Beast, there is something there that wasn’t there before—just wait and see, a few years more.
Questions for authors
- In this preprint, it was noted that libraries that targeted variation in more than 5 codons did not saturate the DNA and protein sequence spaces. Were these libraries synthesised and read? Or is this more of a limit based on a theoretical construct built on the “coupon collector” calculation? Given an even higher throughput technology in synthesising such a library, could this be a direction for future work?
- The ability of the cocktail and HI libraries were found to be able to suppress resistance for about a week. In this series of experiments, were the conditions (such as MOI and duration of experiment) optimised, or might there be room for optimisation of these factors in future work?
- How was the cocktail composition selected—was the selection based on certain phage activity or diversity of the loops represented?
References
[1] Międzybrodzki R, Borysowski J, Weber-Dąbrowska B, Fortuna W, Letkiewicz S, Szufnarowski K, Pawełczyk Z, Rogóż P, Kłak M, Wojtasik E, Górski A (2012) Chapter 3 – Clinical Aspects of Phage Therapy, in Advances in Virus Research (Łobocka M and Szybalski W eds) pp 73-121, Academic Press.
[2] Dedrick RM, Guerrero-Bustamante CA, Garlena RA, Russell DA, Ford K, Harris K, Gilmour KC, Soothill J, Jacobs-Sera D, Schooley RT, Hatfull GF, Spencer H, Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus, Nature Medicine 25(5) (2019) 730-733.
[3] Schooley RT, Biswas B, Gill JJ, Hernandez-Morales A, Lancaster J, Lessor L, Barr JJ, Reed SL, Rohwer F, Benler S, Segall AM, Taplitz R, Smith DM, Kerr K, Kumaraswamy M, Nizet V, Lin L, McCauley MD, Strathdee SA, Benson CA, Pope RK, Leroux BM, Picel AC, Mateczun AJ, Cilwa KE, Regeimbal JM, Estrella LA, Wolfe DM, Henry MS, Quinones J, Salka S, Bishop-Lilly KA, Young R, Hamilton T, Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails To Treat a Patient with a Disseminated Resistant Acinetobacter baumannii Infection, Antimicrobial Agents and Chemotherapy 61(10) (2017) e00954-00917.
Posted on: 27 July 2019 , updated on: 29 September 2019
doi: https://doi.org/10.1242/prelights.12577
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the microbiology category:
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
Digital Microbe: A Genome-Informed Data Integration Framework for Collaborative Research on Emerging Model Organisms
Mixed Alkyl/Aryl Phosphonates Identify Metabolic Serine Hydrolases as Antimalarial Targets
Also in the synthetic biology category:
Discovery and Validation of Context-Dependent Synthetic Mammalian Promoters
Genetically encoded multimeric tags for intracellular protein localisation in cryo-EM
Dissecting aneuploidy phenotypes by constructing Sc2.0 chromosome VII and SCRaMbLEing synthetic disomic yeast
preLists in the microbiology category:
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Also in the synthetic biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Advances in Drug Delivery
Advances in formulation technology or targeted delivery methods that describe or develop the distribution of small molecules or large macromolecules to specific parts of the body.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)