FUS-dependent liquid-liquid phase separation is an early event in double-strand break repair
Preprint posted on 24 August 2020 https://www.biorxiv.org/content/10.1101/798884v2
Article now published in Journal of Cell Biology at http://dx.doi.org/10.1083/jcb.202008030
Categories: biochemistry, cell biology, molecular biology, neuroscience
Context
Liquid-Liquid phase separation (LLPS) coalesces macromolecules from the local cellular milieu to facilitate many biological processes. LLPS has emerged as an important biophysical mechanism to regulate a myriad of the biological process from organizing non-membranous cellular compartments to macromolecules that facilitate cellular and genome function. Notably, in the nucleus, LLPS promotes the spontaneous organization of proteins and nucleic acids with shared functions to facilitate genomic processes like chromatin organization, transcription, RNA processing, DNA replication, and DNA damage response1,2.
In response to DNA damage, cells often commission the proteins available in the local vicinity of the damage for the repair process. This could be the reason for many RNA processing factors to play a crucial role in DNA damage response pathways. Dysregulation of one or many RNA processing factors is known to cause DNA damage and impede genome integrity3. In light of this evidences, the current work investigated the role of LLPS in mediating a DNA/RNA binding protein Fused in sarcoma (FUS) driven DNA damage response.
Key findings
- The investigators made FUS depleted HeLa and SH-SY5Y cells using CRISPR (and shRNA) based technologies to evaluate the role of FUS in the DNA damage response. They found that both cell lines manifested higher levels of DNA damage, as assayed by measuring immunofluorescence staining of DNA damage marker γH2AX. This damage is rescued by ectopic expression of FUS. They also report perturbed DNA damage signaling (assayed by DNA damage marker proteins like γH2AX, 53BP1, pATM, pATR) and low survival rates in FUS depleted cells pharmacologically treated with DNA damaging agents (etoposide and camptothecin).
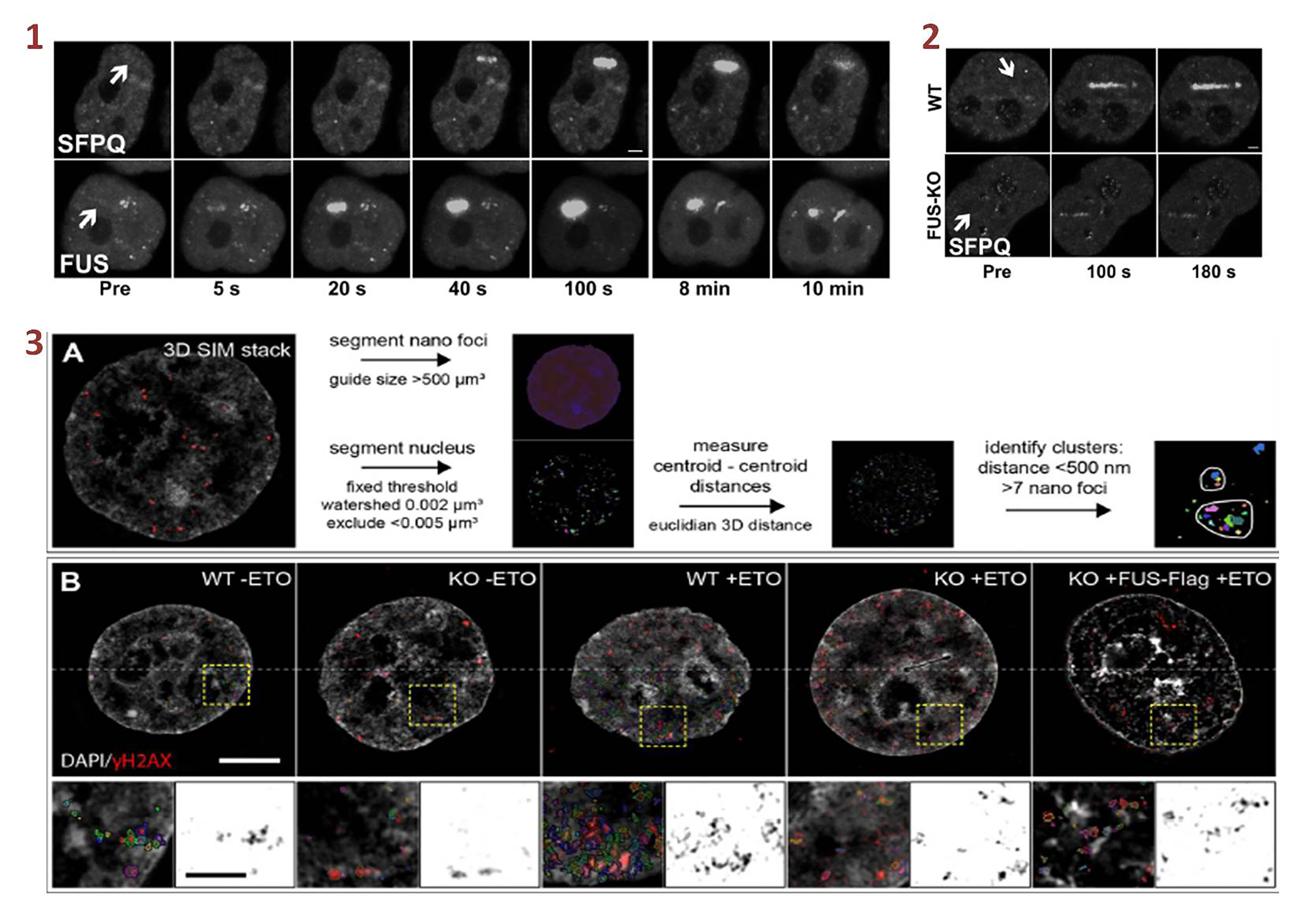
(1) & (2) FUS recruitment to DNA damage sites precedes SFPQ and its absence strongly reduces SFPQ accumulation. (3) FUS is required for γH2AX nano-foci clustering. Taken and modified directly from Levone B et. al., 2020 under a CC-BY 4.0 international license. - Quantification of γH2AX foci or intensities of γH2AX foci is often used as a read-out to measure DNA damage in human cells. However, in earlier work4, the team demonstrated that γH2AX nano-foci could reveal the 3D organization of γH2AX marked nucleosomes at the DNA damaged chromatin. Building on this, they now show that FUS depletion impedes γH2AX nano-foci clustering that can be rescued by ectopic expression of FUS. Thus, they suggest that FUS is necessary for efficient nano-clustering of γH2AX (and other DNA repair factors) at the DNA damaged chromatin.
- To understand the role of FUS in the early DNA damage response, they analyzed the recruitment kinetics of DNA repair factors at laser-induced microirradiated DNA damage sites. They found that FUS was recruited to DNA damage sites that are prominently marked by γH2AX. FUS recruitment to sites of DNA damage was swift (~5s after damage induction) and preceded SFPQ (~20S after damage induction), an interacting protein of FUS. Furthermore, FUS depletion compromised SFPQ recruitment at DNA damage sites. FUS depletion also modified the recruitment dynamics of other DNA repair factors (KU80, NBS1, 53BP1, BRCA1). Thus, they suggest that FUS acts at early time points of DNA damage response.
- Based on earlier studies, the authors hypothesized whether the ability of FUS and SFPQ to phase separate is necessary for their recruitment to damaged DNA. The investigators demonstrate that LLPS acts as an important driver for FUS and SFPQ mediated DNA damage response by evaluating the recruitment kinetics of FUS and SFPQ to DNA damage sites in the presence of LLPS inhibiting chemicals (1,6-hexanediol and ammonium acetate). They also found that γH2AX and 53BP1 foci were reduced in the presence of LLPS inhibiting chemicals. Furthermore, LLPS-compromised FUS variants did not localize to sites of DNA damage and also hindered SFPQ and KU80 recruitment to the DNA damage sites. Thus, they report that LLPS is crucial to recruit DNA repair factors, and FUS-mediated DNA damage response depends on its LLPS nature.
Conclusion and perspective
Previous reports demonstrate the role of FUS, a DNA/RNA binding protein in DNA damage response5. The current study reports that LLPS is an important property of FUS to act at the onset of damaged DNA, and this is an early event for the repair of damaged DNA. Toxic loss of function mutations in multiple RNA processing genes including FUS causes familial amyotrophic lateral sclerosis (ALS). Moreover, cytoplasmic inclusions of FUS protein aggregates were observed in patients with sporadic ALS5,6. If and how phase separation of toxic FUS variants explain the etiology of FUS-ALS is yet to be elucidated.
Acknowledgments: Thanks to Silvia Barabino and all the authors of this work for their support.
References
- https://doi.org/10.1242/jcs.235093
- https://doi.org/10.1016/j.tibs.2020.06.007
- https://doi.org/10.1016/j.molcel.2009.06.021
- https://doi.org/10.1038/ncomms15760
- https://doi.org/10.1073/pnas.1611673113
- https://doi.org/10.1016/j.brainres.2014.10.009
Posted on: 5 February 2021
doi: https://doi.org/10.1242/prelights.25121
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biochemistry category:
Structural basis of respiratory complexes adaptation to cold temperatures
Lens Placode Modulates Extracellular Matrix Formation During Early Eye Development
Generalized Biomolecular Modeling and Design with RoseTTAFold All-Atom
Also in the cell biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Structural basis of respiratory complexes adaptation to cold temperatures
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
Also in the molecular biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Nanos2+ cells give rise to germline and somatic lineages in the sea anemone Nematostella vectensis
Plant plasmodesmata bridges form through ER-driven incomplete cytokinesis
AND
Plasmodesmata act as unconventional membrane contact sites regulating inter-cellular molecular exchange in plants
Also in the neuroscience category:
Transcriptional profiling of human brain cortex identifies novel lncRNA-mediated networks dysregulated in amyotrophic lateral sclerosis
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
Synergistic olfactory processing for social plasticity in desert locusts
preLists in the biochemistry category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
Preprint Peer Review – Biochemistry Course at UFRJ, Brazil
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biochemistry deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the cell biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |
Also in the molecular biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the neuroscience category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |











 (No Ratings Yet)
(No Ratings Yet)