A phospho-regulated ensemble signal motif of α-TAT1 drives dynamic microtubule acetylation
Preprint posted on 23 September 2020 https://www.biorxiv.org/content/10.1101/2020.09.23.310235v1
Article now published in Journal of Cell Biology at http://dx.doi.org/10.1083/jcb.202202100
The Takanari Inoue lab explores the dynamic spatial distribution of the conserved acetyltransferase α-TAT1 in HeLa cells: Spatial shuttling of α-TAT1 between nucleus and cytoplasm regulates levels of acetylated microtubules in the cell.
Selected by Sukriti KapoorCategories: cell biology
Background
Mechanical properties of microtubules can be altered by several post-translational modifications (PTMs). One such well-conserved PTM is acetylation. Acetylated microtubules are more stable, better able to resist bending forces, and therefore, are protected from mechanical stress1,2. Tubulin acetylation contributes to the dynamic nature of microtubule organization and function in specialized processes such as intracellular protein trafficking, cilia motility, cell migration and cell division3.
Acetylation occurs on Lysine 40 position of a-tubulin in the cytoplasm and is catalyzed by α-TAT1, a highly conserved mammalian acetyltransferase2. While the mechanistic insights into the catalytical role of α-TAT1 in promoting microtubule stability has been explored earlier, the regulation of intracellular distribution of the acetyltransferase itself has not been looked at in detail4. This preprint aims to understand the spatial regulation of acetyltransferase, α-TAT1, and its impact on the levels of microtubule acetylation in the cell.
(A)
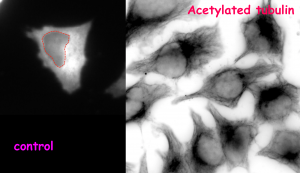
(B)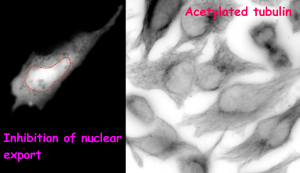
Figure (adapted from the preprint) shows the impact of change in α-TAT1 (acetyltransferase) localization on tubulin acetylation in HeLa cells. In control condition (A), majority of cells show cytoplasmic localization of α-TAT1 (left). Drug based inhibition of nuclear export (B) causes α-TAT1 to be sequestered in the nucleus (left). This leads to a reduction in the levels of acetylated microtubules in the cytoplasm (right).
Key Findings
While the N-terminus of α-TAT1 consists of the catalytic domain, its C -terminus was predicted to be intrinsically disordered and previously less well characterized. In this study, using a combination of sequence prediction and analysis software algorithms, α-TAT1 was predicted to comprise highly conserved regions of both NLS (nuclear localization signal) and NES (nuclear exclusion signal) domains. Upon expressing the fluorescently tagged α-TAT1 protein in HeLa cells, it was found to exhibit a range of localization patterns. Minor fraction of cells showed (a) a nuclear enrichment or (b) a diffuse localization of the protein. In majority of the cells, the protein was found to be exclusively localized to the cytoplasm. This dominant cytosolic distribution was expected since α-TAT1 is known to localize with the acetylated microtubules. Additionally, however, the intracellular fluorescence intensity of the protein was found to vary across time, indicative of a dynamic nature of a-TAT1 localization in cells. This categorical and dynamic localization pattern of a-TAT1 suggested that its function may be regulated by altering its distribution pattern in cells.
How is α-TAT1 localization regulated in the cell? The authors reported an interaction between α-TAT1 and exportin, a member of the nuclear export machinery, which suggested that α-TAT1 may be actively exported out of the nucleus, such that it can catalyze acetylation on microtubules in the cytoplasm. To test this, the authors used a generic drug to inhibit exportin-mediated transport and found that they could effectively trap a large pool of acetyltransferase in the nucleus (Major fraction of the cells showed a diffused localization of the protein). On doing so, expectedly, they were able to drastically reduce the levels of acetylated microtubules in the cytoplasm (animated video). Hence, the spatial switch of protein localization from the cytoplasm to the nucleus or vice versa is an efficient mechanism to regulate its function and alter the acetylation status of microtubules in the cells.
What controls the spatial shuttling of α-TAT1 between the nucleus and the cytoplasm? Upon expressing shorter fragments of the protein in the cells, it was found that the C-terminus of the protein, but not its N-terminus fragment, is preferentially in localized in the cytosol, suggesting that the export is mediated by the C-terminus. Truncation of putative NES domain abrogated its export from the nucleus. However, noticeably, as is the case with drug-mediated inhibition of nuclear export, this mutant protein too exhibited a dominant diffused localization pattern, suggesting that deletion of the NES alone is not sufficient to restrict the protein to the nucleus. Understandably, additional pathways are involved in the regulation of spatial distribution of α-TAT1.
Next, the authors set out to characterize the level of regulation on the putative NLS domain, which is also located on the C-terminus. The predicted NLS domain was found to be bind 14-3-3 proteins, which are reported to promote nuclear exclusion. Since interaction with 14-3-3 proteins conventionally requires phosphorylated serine and threonine residues, the authors subjected the cells to a battery of kinase inhibitors, and found that inhibition of kinases such as CDKs, CK2 and PKA increased the nuclear localization of the protein, such that major fraction of the cells showed diffused α-TAT1 protein localization. α-TAT1, therefore, must be in a phosphorylated state for its efficient nuclear export. Whether this phosphorylation occurs on the predicted NLS domain or not is still unresolved in the current study. [Of note, the current study does not comment on whether the nuclear enrichment observed upon kinase inhibition is because of a reduced affinity of 14-3-3 proteins for α-TAT1 or whether these kinases regulate α-TAT1 localization through an independent alternative mechanism. Further, the significance of interaction of α-TAT1 with 14-3-3 proteins on its spatial distribution pattern or on the regulation of tubulin acetylation has not been validated experimentally in the current version of the study.]
On further exploring the theme of phospho-regulation of α-TAT1 spatial distribution, the binding of the predicted NLS domain to importin, a member of the nuclear import machinery, was disrupted by mutating the basic residues flanking the NLS to neutral charged amino acid, alanine. This was done because importin has been reported to preferentially interact with basic residues enriched in the NLS domain. These mutations resulted in enhanced nuclear enrichment of α-TAT1, indicating that phosphorylation on these particular residues might be important for mediating nuclear export/import. [An important caveat of the experiment is that the whether these mutations indeed disrupt the interaction with importin or 14-3-3 proteins has not been tested in this study.]
In summary, this preprint explores the key processes by which the spatial distribution of acetyltransferase, α-TAT1, can be modulated in the cell. Nuclear export to the cytoplasm is independently carried out through the NES and NLS domains: – (a) direct binding of the NES domain to nuclear export machinery and (b) phosphorylation on the NLS domain.
What I like about this preprint
This preprint has exploited a combination of sequence prediction tools and bioinformatic analysis to experimentally characterize the unique spatial regulation of α-TAT1. Abnormal tubulin acetylation levels have been reported in several pathological conditions, such as neurodegenerative disorders and cancer3. The current study has provided a new outlook on exploring microtubule acetylation and associated mechanical properties of the cell with respect to the dynamic spatial distribution of the acetyltransferase, α-TAT1. While the precise mechanism of regulation of α-TAT1 distribution remains unclear from this study, it would be interesting to see how the delicate balance of kinases and phosphatases in the cell determines the spatial distribution of α-TAT1, and consequently microtubule acetylation and mechanical behaviour of the cell.
Future prospects and questions for the authors
- Could the authors please describe or speculate on a scenario in which the change in microtubule acetylation, due to sequestration of α-TAT1 in the nucleus, directly modulates a cell biological process/ behaviour?
- Could the authors comment on the time it would take for the spatial switch in distribution of α-TAT1, i.e., from the nucleus to the cytoplasm or from the cytoplasm to the nucleus, so as to significantly alter microtubule acetylation status of the cell? How much time would it take for cells to elicit any meaningful response to changes in microtubule acetylation?
- The acetylation status of microtubules has been reported to change in certain neurodegenerative disorders and may also be important for biological processes such as migration of cancer cells. Could the authors predict the spatial distribution pattern of α-TAT1 in such conditions?
- What do the authors have to say about the significance of kinases they found in their screen and their impact on microtubule acetylation and eventually, cell function? Additionally, the kinase inhibition screen in the study was done over several hours to overnight. Do the authors think that a shorter window of kinase inhibition could drive a meaningful, fast-acting response in the cell?
- Here, 14-3-3 proteins have been shown to interact with α-TAT1. However, in the current study, it is not immediately clear that 14-3-3 proteins might directly affect the phosphorylation status of α-TAT1 or in fact, play any role in its intracellular spatial distribution. Can the authors explain briefly why they think 14-3-3 proteins would be an important future target for studying microtubule acetylation?
- Microtubule acetylation may be important for the formation of specialized microtubule structures, such as the mitotic spindle. Further, α-TAT1 co-localizes with the microtubules at the time of formation of spindle. During mitosis, there is breakdown of the nuclear envelope, and therefore, should one not expect to see any important role of the NES and NLS domains and their potential phosphor-regulation identified in the study, during cell division?
- This paper has helped characterize key processes and proteins that regulate the spatial distribution of α-TAT1 in microtubule acetylation. Could the authors briefly comment on whether microtubule de-acetyltransferases (say, histone de-acetylase) could be governed by a similar process, if at all?
References
- Hammond, J. W., Cai, D. & Verhey, K. J. Tubulin modifications and their cellular functions. Curr. Opin. Cell Biol. 20, 71–76 (2008).
- Janke, C. & Montagnac, G. Causes and Consequences of Microtubule Acetylation. Curr. Biol. 27, R1287–R1292 (2017).
- Nekooki-Machida, Y. & Hagiwara, H. Role of tubulin acetylation in cellular functions and diseases. Med. Mol. Morphol. 53, 191–197 (2020).
- Kalebic, N. et al. Tubulin Acetyltransferase αTAT1 Destabilizes Microtubules Independently of Its Acetylation Activity. Mol. Cell. Biol. 33, 1114–1123 (2013).
Posted on: 2 December 2020
doi: https://doi.org/10.1242/prelights.26003
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cell biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Structural basis of respiratory complexes adaptation to cold temperatures
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
preLists in the cell biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |











 (No Ratings Yet)
(No Ratings Yet)