Maintenance of spatial gene expression by Polycomb-mediated repression after formation of a vertebrate body plan
Preprint posted on 12 November 2018 https://www.biorxiv.org/content/early/2018/11/12/468769
Article now published in Development at http://dx.doi.org/10.1242/dev.178590
Though ezh2-mutant zebrafish form grossly normal body plans, key developmental factors are dysregulated early on, which dooms the embryo to later failure of organogenesis and lethality.
Selected by Yen-Chung ChenCategories: cell biology, developmental biology, genomics
Background and context
Polycomb repressive complex 2 (PRC2) mediates methylation of lysine 27 on histone 3 (H3K27me3). H3K27me3 is a chromatin mark associated with gene repression, and the disruption of PRC2 results in grave disruption of development. In flies, body segments are organized wrongly when H3K27me3 is disrupted, and mouse embryos die before gastrulation, presumably as a result of failed differentiation. These severe consequences have made H3K27me3 one of the best studied histone modifications in development, and several critical genes governing segmentation or fate specification have been shown to be under the control of PRC2 via H3K27me3 [Reviewed in 1 and 2].
Because mammalian embryos fail to gastrulate without PRC2, the function of PRC2 in development is mostly studied by differentiating stem cells in vitro. Although the in vivo functions of PRC2 are well characterized in flies, the mode of action of Drosophila PRC2 is starkly different from vertebrate PRC2. In a previous study from the Kamminga lab, they used a germ cell transplantation technique in zebrafish to generate a PRC2 loss-of-function model by mutating ezh2, a core component of PRC2. They found out that even without ezh2, the fish remains grossly normal through segmentation until organogenesis [3]. The power of this model enables Rougot et al. to investigate the consequences of the absence of H3K27me3 in gene regulation in vivo in the current preprint.
Key findings
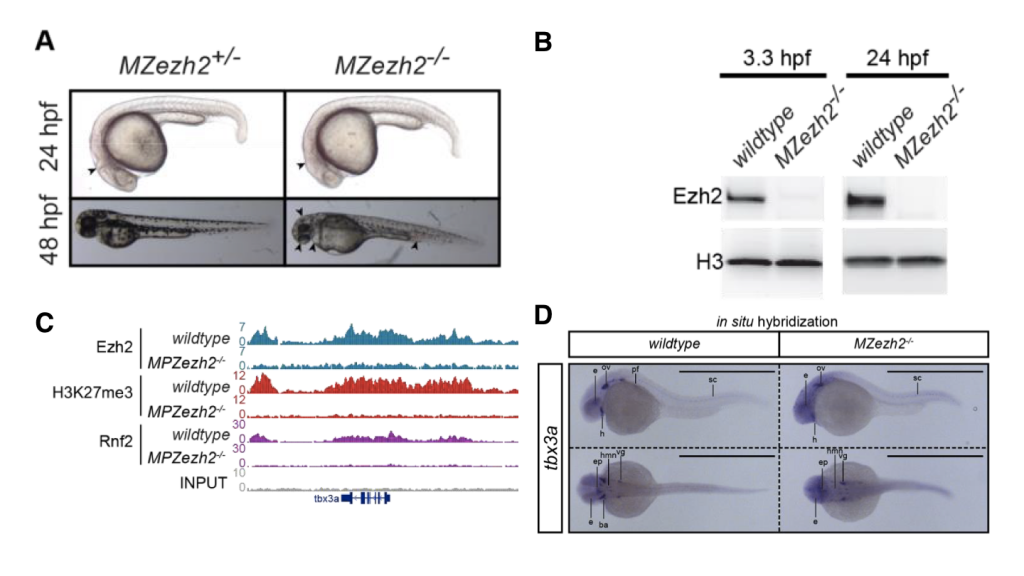
(A) ezh2-mutant embryo develops largely normal up to 48 hours post-fertilization (from Figure 1A)
(B) ezh2-mutants are steadily deprived of ezh2 during early embryogenesis (from Figure 1B).
(C) The landscape of histone marks and the binding of writers are perturbed (from Figure 1G).
(D) Spatial expression of histone mark-regulated genes is altered in the absence of ezh2 (from Figure 6A).
Rougot et al. showed that in maternal-zygotic ezh2mutant zebrafish, the level of H3K27me3 decreases to a level barely detectable with ChIP-seq. Interestingly, besides the absence of H3K27me3, rnf2, the core component of PRC1, failed to find its normal targets in ezh2 mutants, though the subunits of PRC1 are present in ezh2mutants. It has been unclear whether the action of PRC1 is dependent on the disposition of H3K27me3, and the current study provides a new line of evidence that in the early stages of zebrafish development, PRC1 requires functional PRC2 to work properly.
In the absence of H3K27me3, H3K4me3, a histone modification associated with transcriptionally active state, is enriched in otherwise H3K27me3-occupied gene loci. Consistent with the important role H3K27me3 plays in developmental gene regulation, genes regulating developmental processes gain H3K4me3 and are upregulated in the absence of H3K27me3. The authors further performed mass spectrometry to check if these epigenetic and transcriptomic changes resulted in altered proteome. Indeed, the proteomic study shows an up-regulation of both a subset of genes occupied by H3K27me3 in the wildtype and the genes gaining H3K4me3 in the ezh2 mutant.
To further examine segment-specific effects of H3K27me3 disruption, the authors first performed RT-qPCR for different body segments to evaluate the expression pattern of hox genes, which are expressed with a clear segmental pattern and govern the head-to-tail body plane. In ezh2 mutants, thoracic hox genes (e.g. hoxa9a and hoxa9b) are up-regulated in the rostral part of body. More caudal hox genes (e.g. hoxa11b and hoxa13b) are also up-regulated but to a lesser extent. These caudal hoxgenes might be repressed by other mechanisms in addition to H3K27me3. Then, the authors performed in situhybridization of several transcription factors with known spatial expression patterns, including the tbx family, isl1, and gsc. Ectopic expression of every transcription factor examined in mutants. The most extreme change is observed in gsc. The normal expression pattern for gsc is confined in telencephalon and diencephalon, branchial arches, and otic vesicle, while in ezh2 mutants, the expression becomes ubiquitous despite being weak.
In short, Rougot et al. used ezh2 mutant zebrafish to show that the early developmental process is robust against the absence of H3K27me3 and proceeds in a grossly normal pattern without H3K27me3. Nonetheless, dysregulation of expression of developmental regulators is detected at the RNA and protein level at 24 hpf, and the spatial patterns of these developmental regulators are perturbed to various extent without H3K27me3.
Why I like this preprint
Histone modifications have long been proposed as an important factor in development to ensure precise coordination of differentiation processes. Indeed, mechanistic studies corroborate the roles of histone modifications in gene regulation and identify many processes they are involved in. Nevertheless, many studies on histone modification-mediated regulation in mammals are performed in vitro, and phenotypic and mechanistic discrepancies between in vivo and in vitromodels are not uncommon. Besides, although the well-defined and scalable nature of cell models enables detailed characterization of the mechanism and a great variety of manipulations, the lack of higher order organization and interactions between different cell types in cell models poses a great challenge for researchers hoping to assess the crosstalk between different regulatory mechanisms. For example, it is common that a striking mutant phenotype in cell models is less severe and presumably compensated in vivobecause environmental cues might provide information that compensates the dysfunction. Thus, it is an invaluable resource to have a vertebrate model of PRC2 loss of function, and this study shows the degree of erosion of spatial expression pattern differs a lot among the genes examined, which might suggest H3K27me3 contributes differently to the repression of H3K27me3-marked genes.
Open questions
- H3K27me3 is reported to safeguard cell identities. Different cell identities are often controlled by antagonizing developmental cues and undergo transcriptomic changes in opposite direction. Would it be possible to examine each germ layer individually to see if the cells gain more plasticity or dual identity in the absence of H3K27me3? This could also rule out the possibility that the transcriptomic changes in different germ layers of ezh2mutant cancel out each other when an embryo is profiled as a whole.
- It is still unclear how PRC2 finds its target loci to tri-methylate. Would it be feasible to re-introduce ezh2 later in development in an ezh2mutant zebrafish to see if H3K27me3 at certain loci could be established properly to see whether the guiding signal that leads PRC2 to its target loci is transient or not?
References
- Kassis, J. A., Kennison, J. A. & Tamkun, J. W. Polycomb and Trithorax Group Genes in Drosophila. Genetics. 206,1699–1725 (2017).
- Aloia, L., Di Stefano, B. & Di Croce, L. Polycomb complexes in stem cells and embryonic development. Development 140, 2525 LP-2534 (2013).
- San, B. et al.Normal formation of a vertebrate body plan and loss of tissue maintenance in the absence of ezh2. Rep. 6,24658 (2016).
Posted on: 7 January 2019 , updated on: 18 January 2019
doi: https://doi.org/10.1242/prelights.6996
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cell biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Structural basis of respiratory complexes adaptation to cold temperatures
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
Also in the developmental biology category:
Temporal constraints on enhancer usage shape the regulation of limb gene transcription
OGT prevents DNA demethylation and suppresses the expression of transposable elements in heterochromatin by restraining TET activity genome-wide
Actin polymerization drives lumen formation in a human epiblast model
Also in the genomics category:
Temporal constraints on enhancer usage shape the regulation of limb gene transcription
Transcriptional profiling of human brain cortex identifies novel lncRNA-mediated networks dysregulated in amyotrophic lateral sclerosis
A long non-coding RNA at the cortex locus controls adaptive colouration in butterflies
AND
The ivory lncRNA regulates seasonal color patterns in buckeye butterflies
AND
A micro-RNA drives a 100-million-year adaptive evolution of melanic patterns in butterflies and moths
preLists in the cell biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |
Also in the developmental biology category:
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the genomics category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |











 (No Ratings Yet)
(No Ratings Yet)