Metabolic dissimilarity determines the establishment of cross-feeding interactions in bacteria
Preprint posted on 10 October 2020 https://www.biorxiv.org/content/10.1101/2020.10.09.333336v1
Article now published in Current Biology at http://dx.doi.org/10.1016/j.cub.2021.10.019
Don’t say ‘no’ to strangers: Cross-feeding relationships between bacteria are driven by metabolic distance
Selected by Hiral ShahCategories: ecology, microbiology
Introduction
Microbial consortia are mixed microbial communities that may be constructed synthetically or derived from natural environments. Microbial consortia impact a variety of environmental, agricultural, and medical applications including bioremediation, polymer degradation, crop improvement, synthesis of nutrition supplements, and biofilm formation (1,2). However, the complex structure and dynamic nature of such communities makes designing and establishing potent microbial consortia far from trivial (3). A key step forward is to better understand the rules that determine the evolution of microbial interactions within these communities (4). A large proportion of microbial interactions rely on an exchange of essential metabolites between two or more bacterial partners. Many of those cross-feeding interactions are unidirectional interactions, meaning that one bacterium utilizes a metabolite that is provided by another one (5). Examples for this include auxotrophic bacteria, which benefit by utilizing a metabolite (eg. an amino acid) that is provided by a prototropic neighbor. In such cases, the benefit is often asymmetrically distributed, with the prototroph bearing the cost of metabolite biosynthesis and release. While cross-feeding is ubiquitous, it remains unclear how these relationships are established.
The authors discuss two possible hypotheses for how these interactions may occur. First, the similarity hypothesis suggests that microbial cultures support the growth of close relatives diverting resources to kin. However, microbial communities are often heterogeneous involving very diverse bacterial groups. On the contrary, the dissimilarity hypothesis expects cross-feeding interactions should be more common amongst genetically more distantly-related organisms, thus favoring less similar donor-recipient pairs to establish cross-feeding interactions. To distinguish between these two possibilities, the authors study cross-feeding between phylogenetically diverse auxotroph-prototroph bacterial pairs.
Key Findings
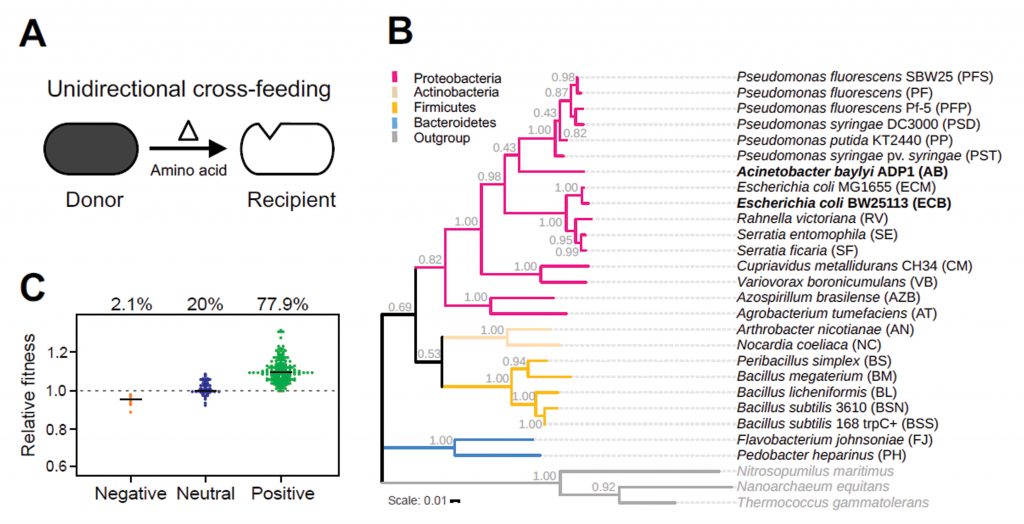
What determines the establishment of these cross-feeding interactions? The authors address this question using a range of different bacterial species as potential amino acid donors to strains of Escherichia coli and Acinetobacter baylyi that were auxotrophic for the two amino acids histidine and tryptophan. Prototrophic genotypes belonged to diverse phylogenetic groups closely and distantly related to the focal recipients. The selected donors lacked any previous coevolutionary history with the recipient, thus ruling out a pre-existing interaction other than the one being investigated. First, the growth of recipients in coculture with donors was compared to the growth they achieved in monoculture. 78% of instances showed a positive response to the presence of the donor (Fig. 1A), suggesting that cross-feeding relationships are easily established between donor and recipient.
In order to define metabolites supporting the growth benefit, donor culture supernatants were analyzed. Interestingly, the growth benefit was proportional to both the total amino acid concentration in addition to the concentration of the focal amino acid suggesting that the recipient may utilize other freely available amino acids apart from just the required one. Next the authors asked whether the difference in amino acid profiles could explain the differences in growth benefits provided by different donor groups.
Could the different amino acid profiles of these diverse donors provide insight into which of the two hypotheses was at play? It turns out that the more different the amino acid profiles produced by donor genotypes were, the higher the growth benefit it provided to the recipient, thus supporting the dissimilarity hypothesis. Furthermore, the growth benefit of recipients also correlated positively with the phylogenetic distance to donors. With different amino acid profiles already favoring growth, was phylogenetic distance only a proxy for metabolic distance? Addressing this possibility, the authors next compared the metabolic networks between different donors. Once again growth benefit was directly proportional to metabolic distance of the donor-recipient pair. Thus, overall distances favored cross-feeding interactions.
Can these findings be extended to many more different microbial interactions or does it apply to just this subset analyzed? In silico simulation of over 330,000 pairwise interactions between 818 bacteria commonly found in the gut showed that the propensity of metabolic exchange follows a saturation curve in response to metabolic distance, once again supporting dissimilarity as a key determinant driving cross-feeding relationships.
References
- Zhang, S., Merino, N., Okamoto, A. & Gedalanga, P. Interkingdom microbial consortia mechanisms to guide biotechnological applications. Microb. Biotechnol. 11, 833–847 (2018). doi.org/10.1111/1751-7915.13300
- Qian, X. et al. Biotechnological potential and applications of microbial consortia. Biotechnol. Adv. 40, 107500 (2020). doi.org/10.1016/j.biotechadv.2019.107500
- Kong, W., Meldgin, D. R., Collins, J. J. & Lu, T. Designing microbial consortia with defined social interactions. Nat. Chem. Biol. 14, 821–829 (2018). doi.org/10.1038/s41589-018-0091-7
- Giri, S., Shitut, S. & Kost, C. Harnessing ecological and evolutionary principles to guide the design of microbial production consortia. Curr. Opin. Biotechnol. 62, 228–238 (2020). 10.1016/j.copbio.2019.12.012
- D’Souza, G. et al. Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat. Prod. Rep. 35, 455–488 (2018). https://doi.org/10.1039/C8NP00009C
Posted on: 23 October 2020
doi: https://doi.org/10.1242/prelights.25435
Read preprintHave your say
Sign up to customise the site to your preferences and to receive alerts
Register hereAlso in the ecology category:
How the liver contributes to stomach warming in the endothermic white shark Carcharodon carcharias
Pervasive sublethal effects of agrochemicals as contributing factors to insect decline
Multimodal interactions in Stomoxys navigation reveals synergy between olfaction and vision
Also in the microbiology category:
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
Digital Microbe: A Genome-Informed Data Integration Framework for Collaborative Research on Emerging Model Organisms
Mixed Alkyl/Aryl Phosphonates Identify Metabolic Serine Hydrolases as Antimalarial Targets
preLists in the ecology category:
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
Bats
A list of preprints dealing with the ecology, evolution and behavior of bats
| List by | Baheerathan Murugavel |
Also in the microbiology category:
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)
4 years
Christian Kost
Thank you very much Hiral for summarizing our work in this very nice piece!