MicroRNA-mediated control of developmental lymphangiogenesis
Preprint posted on 20 February 2019 https://www.biorxiv.org/content/10.1101/555664v1
Article now published in eLife at http://dx.doi.org/10.7554/eLife.46007
microRNA, macro responsibilities: miRNA-204-mediated regulation of Nfatc1 is important for development of proper lymphatic vessels.
Selected by Rudra Nayan DasCategories: developmental biology, molecular biology
Background
Lymphatic vessels are crucial components in vertebrate fluid homeostasis. They build a network of vessels and nodes that is important for drainage of interstitial fluid, circulation of immune cells and transport of dietary lipids.
Over the years, studies on the molecular mechanisms that control lymphatic development have revealed the involvement of several signalling pathways and transcriptional regulators. However, information on post-transcriptional regulation of lymphatic development is limited. MicroRNAs (miRNAs) are non-coding, ~20-24 nucleotide sequences that regulate gene expression by silencing the mRNAs of their target genes. Post-transcriptional regulation through specific miRNAs has been documented to be important for normal development of a variety of tissues and as many as 60% of human protein-coding genes are predicted to have miRNA target sites1. In the context of lymphatic development, some miRNAs have been reported to have a role in lymphatic endothelial cell (LEC) specification, development and inflammation2. However, only a few of these studies have utilized in vivo model systems.
In the past decade, zebrafish has been well-established as a model system for lymphatic development3,4. In this study, Jung et al. combined human endothelial cell culture system and zebrafish embryos to identify how miRNA-204 and its target nfatc1 is required to generate optimal levels of lymphatic structures.
Important results
To identify miRNAs that are enriched in lymphatic endothelial cells (LECs), Jung et al. performed small RNA sequencing on human dermal lymphatic microvascular endothelial cells (HMVEC-dLy, representing LECs) and human umbilical vein endothelial cells (HUVECs, representing blood endothelial cells). Comparative analysis of these datasets revealed 98 differentially expressed miRNAs, among which 30 were highly expressed in LECs. miR-204, with 105 times higher expression in LECs, was singled out for in vivo characterization.
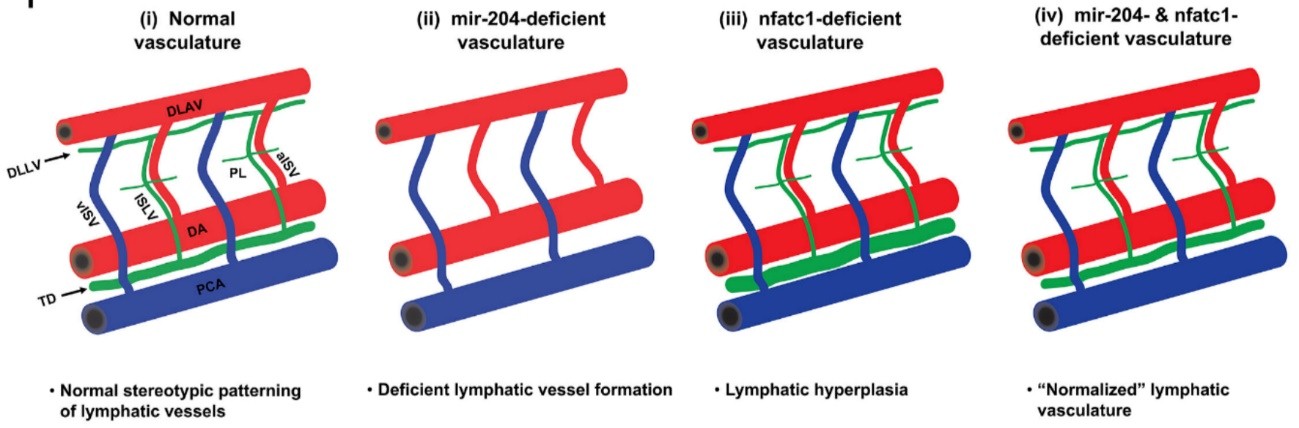
Enrichment of miR-204 was also found in developing LECs of zebrafish embryos, but the zebrafish genome harbors three paralogues of miR-204 (referred as miR-204-1, 204-2 and 204-3), all of which produce a mature miR-204 whose sequence that is 100% identical to its human counterpart. Injection of a pan-miR-204 morpholino, that suppresses the mature miR-204 produced from all the three loci, caused loss of early lymphatic structures in the trunk of the embryos. However, a CRISPR mutant that was designed to disrupt 204-1 showed no phenotype and caused only 20% reduction in the total miR-204 content, indicating that 204-2 and/or 204-3 can compensate for this mutation. Using morpholinos that specifically suppressed miR-204-1, 204-2, 204-3 or a combination of them, the authors could conclude that miR-204-1 and 204-2 are the major contributors towards the total miR-204 and suppressing products from these two loci together caused prominent loss of early lymphatics in the trunk. The role of miR-204 as a positive regulator for lymphatic formation was further strengthened when lymphatic-specific overexpression of miR-204 resulted in faster development of the thoracic duct, the early forming lymphatic vessel in the trunk.
To identify the gene(s) regulated by miR-204, the authors utilized RNA22, a computational tool for miRNA target discovery5, and identified nfatc1 as a possible target. The authors showed that miR-204 can indeed supress nfatc1 and it is mediated through miR-204 binding in the 3’UTR region of the nfatc1 transcript. Furthermore, suppression of miR-204 indeed increased endogenous nfatc1 transcript levels both in human LECs and in zebrafish. Loss of Nfatc1 has been previously shown to cause lymphatic hyperplasia in mouse6. Similarly, Jung et al. found thoracic duct enlargement upon suppression of nfatc1 transcript or inhibition of Nfatc1 downstream pathway.
Thus, while miR-204 promotes lymphatic growth, Nfatc1 seems to restrict lymphatic growth (since its downregulation causes lymphatic hyperplasia). To establish a functional link between these two, morpholino-mediated co-suppression of miR-204 and nfatc1 was performed. As expected, the downregulation of both ended up giving rise to a somewhat normal lymphatic vasculature.
Why I chose this preprint
In this work, Jung et al. have successfully utilized human cells and zebrafish in vivo system to identify an evolutionary conserved post-transcriptional regulatory mechanism for lymphatic development. Apart from presenting a new regulator of lymphatic development, this study also provides strong evidence for lymphatic developmental conservation between mammalian and fish model systems.
There is also something exciting about identifying miRNAs that strongly influence certain biological processes. Since miRNAs are small sequences, they can be easily delivered, sometimes with suitable modifications, for miRNA-based therapeutics7. In fact, several miRNAs have been utilized for preclinical studies, and a few are being considered for clinical trials. This is of importance in the context of lymphatics, as lymphatic dysfunction causes a range of debilitating conditions, called lymphedema, which lacks a proper cure. Identification of miRNAs that can have impact on lymphatic development and maintenance offers opportunities for therapeutics, and thus this study, although very preliminary, opens such an opportunity.
Questions to the authors
- The data presented in this manuscript provides support for a model where Nfatc1 is suppressing LEC proliferation, and miR-204, by suppressing the levels of Nfatc1, allows for optimal lymphatic development. However, the presence of Nfatc1 indicates some role for it either in the early lymphatic progenitors or in the PCV cells (that generate lymphatic progenitors). Have the authors attempted to detect Nfatc1 (using in situ hybridization or in the transcriptome of sorted cells) in the zebrafish PCV/early LECs? It would be great if the authors can share their views on the possible role of Nfatc1 in this context.
- While there is a clear evidence of miR-204 suppressing Nfatc1, it is still possible that miR-204 regulates other transcripts required for lymphatic development (as also discussed by the authors). Do the authors think that a Nfatc1 overexpression experiment can resolve this issue based on whether it phenocopies miR-204 suppression? Have the authors also identified, in the miRNA target prediction, any other molecular players in the lymphatic pathway as potential targets of miR-204?
- There are number of reports of involvement of miR-204 in certain cancers, where miR-204 is described as a tumor suppressor. Other unrelated studies have also implicated Nfatc1 in certain cancers and in maintenance of stem cell quiescence8. Do the findings from these studies provide any interesting clues that can explain some of the phenotypes described in the present work?
- This work utilizes morpholinos for many of the important experiments. In recent years, morpholino usage in zebrafish has been recommended with caution9. I feel it would be great for other zebrafish researchers if the authors share some of the crucial aspects of their experiments that allows for a greater reliability in their morpholino experiments. I was also wondering if the authors plan to use a double mutant for 204-1 and 204-2 for a more precise demonstration of their findings.
References
- Gebert, L. F. R. & MacRae, I. J. Regulation of microRNA function in animals. Nature Reviews Molecular Cell Biology (2019). doi:10.1038/s41580-018-0045-7
- Yee, D., Coles, M. C. & Lagos, D. microRNAs in the lymphatic endothelium: Master regulators of lineage plasticity and inflammation. Frontiers in Immunology (2017). doi:10.3389/fimmu.2017.00104
- Yaniv, K. et al. Live imaging of lymphatic development in the zebrafish. Nat. Med. 12, 711–716 (2006).
- Hogan, B. M. & Schulte-Merker, S. How to Plumb a Pisces: Understanding Vascular Development and Disease Using Zebrafish Embryos. Developmental Cell (2017). doi:10.1016/j.devcel.2017.08.015
- Miranda, K. C. et al. A Pattern-Based Method for the Identification of MicroRNA Binding Sites and Their Corresponding Heteroduplexes. Cell (2006). doi:10.1016/j.cell.2006.07.031
- Norrmén, C. et al. FOXC2 controls formation and maturation of lymphatic collecting vessels through cooperation with NFATc1. J. Cell Biol. (2009). doi:10.1083/jcb.200901104
- Rupaimoole, R. & Slack, F. J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nature Reviews Drug Discovery (2017). doi:10.1038/nrd.2016.246
- Horsley, V., Aliprantis, A. O., Polak, L., Glimcher, L. H. & Fuchs, E. NFATc1 Balances Quiescence and Proliferation of Skin Stem Cells. Cell (2008). doi:10.1016/j.cell.2007.11.047
- Stainier, D. Y. R. et al. Guidelines for morpholino use in zebrafish. PLoS Genet. (2017). doi:10.1371/journal.pgen.1007000
Posted on: 7 March 2019
doi: https://doi.org/10.1242/prelights.9240
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the developmental biology category:
Temporal constraints on enhancer usage shape the regulation of limb gene transcription
OGT prevents DNA demethylation and suppresses the expression of transposable elements in heterochromatin by restraining TET activity genome-wide
Actin polymerization drives lumen formation in a human epiblast model
Also in the molecular biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Nanos2+ cells give rise to germline and somatic lineages in the sea anemone Nematostella vectensis
Plant plasmodesmata bridges form through ER-driven incomplete cytokinesis
AND
Plasmodesmata act as unconventional membrane contact sites regulating inter-cellular molecular exchange in plants
preLists in the developmental biology category:
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the molecular biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (1 votes)
(1 votes)