Mutational signatures are jointly shaped by DNA damage and repair
Preprint posted on 11 February 2020 https://www.biorxiv.org/content/10.1101/686295v2
Article now published in Nature Communications at http://dx.doi.org/10.1038/s41467-020-15912-7
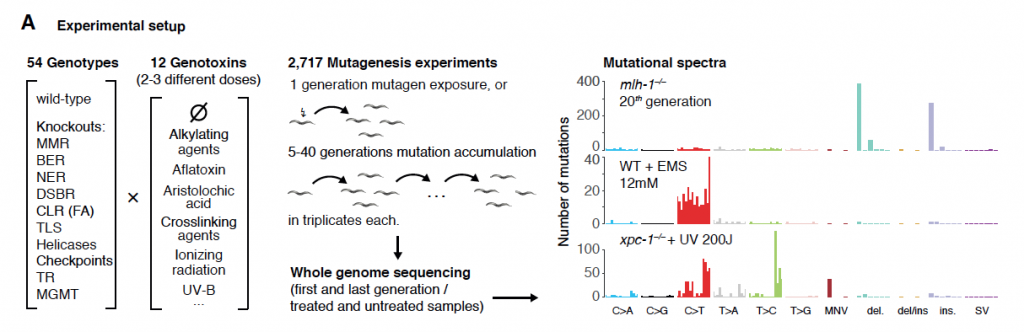
A combinatory mutational screen in C. elegans, combining 54 DNA repair mutants and 12 genotoxins, demonstrates that mutations are a result of both DNA damage and failed repair.
Selected by Kerryn ElliottCategories: cancer biology, cell biology, genetics, genomics
Background
The DNA of a given cell is constantly altered by DNA replication errors and genotoxic stresses. An assemblage of repair pathways specifically designed to keep mutations at bay usually efficiently repairs these alterations. Occasionally this process fails, and DNA lesions escape detection, or are repaired by error-prone pathways, leading to diseases such as cancer.
With the recent advances in whole genome sequencing, the mutational spectra of human cancers have been dissected into individual mutagenic processes using computational pattern recognition programs that correlate genotoxic exposures, such as UV light, or DNA repair deficiencies, such as defects in mismatch repair (MMR), with certain mutational signatures. There are now more than 50 mutational signatures identified computationally, and approximately one third of these have no known etiology (Alexandrov et. al., 2020). It is possible that these unknown signatures represent combinations of mutagens and repair pathway defects. In this paper the authors perform a combinatorial mutagenesis screen with 12 genotoxic agents on 53 different DNA repair mutants in C. elegans to show that mutations are a result of both DNA damage AND failed repair.
Main findings
The combination of using genotoxin treatment in backgrounds of DNA repair deficiency was a clever approach to understand whether the genotoxins themselves, or the failed repair contributed more to the mutational signature. The authors found that over 40% of the combination treatments demonstrated unique mutational signatures or altered mutation rates, but unexpectedly there were also combinations where the loss of translesion synthesis enzymes (TLS) led to a decrease in mutagenesis. This suggests that the majority of mutations induced by genotoxins are caused by error-prone translesion synthesis. By comparing to the genotoxin treatments on a wild type background, the authors were able to attribute the mutations to either the genotoxin itself (54%), DNA repair deficiencies themselves (26%), and to positive (23%) and negative (3%) genotoxin-repair interactions. Notably, nucleotide excision repair (NER) was found to be responsible for the majority of the repair following genotoxin exposure, as the loss of NER proteins increased the mutational load substantially.
Interestingly, while the interactions between genotoxins and DNA repair deficiencies usually retained a similar mutation spectrum with an increased mutation rate, around 10% of the interactions resulted in a change in the mutational spectrum. The authors found lesion-specific mutations for alkylating agents under different DNA repair deficiency backgrounds. The alkylating agent MMS was highlighted, as MMS can alkylate A and G nucleotides to generate different modifications, mainly N3-methyladenine and O6-methylguanine. These modifications are processed by different repair pathways, and therefore blocking one or the other pathway led to a different mutational outcome. Using the specific mutants, the authors were able to link the translesion synthesis enzyme Pol κ to the repair of specific adenine lesions (N3-methyladenine), and the alkyl-transferase AGT-1 to the repair of guanine modifications (O6-methylguanine). They also proposed that the error-prone TLS enzyme Pol ζ was responsible for a large proportion of the additional mutations occurring in the knockouts, as the Pol ζ deficient strain showed a decrease in single base mutations, and instead showed an increase in large deletions, possibly due to fork-stalling. It is this combination of DNA repair deficiency alongside genotoxin exposure which makes this paper a very interesting read!
Why I chose this paper:
Mutational signatures, particularly those driven by the combination of damage and repair are a main research interest of mine. The use of C. elegans as a model organism allowed the combination of treatments to be studied in detail, leading to the ability to dissect out the cause of the mutational signature, be it the mutagen itself or the associated repair process.
This work is important for the field to understand the heterogeneity of mutagenesis and the importance of the combination of damage and failed repair in generating mutations.
Questions to the authors:
Are these results directly relatable to human cancers, given the treatment was on C. elegans? You mention an organism-specific spectrum in Cisplatin, but are there additional complexities (transcription factor families, orthologs, levels of mutagen exposure in human life etc.) that are missing in the model organism that may influence mutations in human cells?
It is interesting that some of these “created” signatures can be detected in human cancers. How do you propose we move forward to understand “combinatory” signatures in humans? How confident are you in the current mutational signature spectra, particularly in assigning etiology?
The increase in large deletions and decrease in SNVs in some TLS mutants was quite interesting to me. Could you expand on your reasoning as to why this might happen, as you suggested this was likely due to replication stalling and fork collapse? Has this been detected in humans, or are there other TLS enzymes that take over this role and prevent large deletions from occurring?
References:
Alexandrov, L.B., Kim, J., Haradhvala, N.J. et al. The repertoire of mutational signatures in human cancer. Nature 578, 94–101 (2020).
Posted on: 24 February 2020 , updated on: 25 February 2020
doi: https://doi.org/10.1242/prelights.16938
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cancer biology category:
Spatial transcriptomics elucidates medulla niche supporting germinal center response in myasthenia gravis thymoma
Invasion of glioma cells through confined space requires membrane tension regulation and mechano-electrical coupling via Plexin-B2
Modeling transcriptional profiles of gene perturbation with a deep neural network
Also in the cell biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Structural basis of respiratory complexes adaptation to cold temperatures
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
Also in the genetics category:
Temporal constraints on enhancer usage shape the regulation of limb gene transcription
A long non-coding RNA at the cortex locus controls adaptive colouration in butterflies
AND
The ivory lncRNA regulates seasonal color patterns in buckeye butterflies
AND
A micro-RNA drives a 100-million-year adaptive evolution of melanic patterns in butterflies and moths
A revised single-cell transcriptomic atlas of Xenopus embryo reveals new differentiation dynamics
Also in the genomics category:
Temporal constraints on enhancer usage shape the regulation of limb gene transcription
Transcriptional profiling of human brain cortex identifies novel lncRNA-mediated networks dysregulated in amyotrophic lateral sclerosis
A long non-coding RNA at the cortex locus controls adaptive colouration in butterflies
AND
The ivory lncRNA regulates seasonal color patterns in buckeye butterflies
AND
A micro-RNA drives a 100-million-year adaptive evolution of melanic patterns in butterflies and moths
preLists in the cancer biology category:
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Anticancer agents: Discovery and clinical use
Preprints that describe the discovery of anticancer agents and their clinical use. Includes both small molecules and macromolecules like biologics.
| List by | Zhang-He Goh |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
Also in the cell biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |
Also in the genetics category:
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the genomics category:
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |











 (No Ratings Yet)
(No Ratings Yet)