Optogenetic control of Wnt signaling for modeling early embryogenic patterning with human pluripotent stem cells
Preprint posted on 10 June 2019 https://www.biorxiv.org/content/10.1101/665695v1
Article now published in Development at http://dx.doi.org/10.1242/dev.201386
Stem cells see the light! Controlling WNT signaling using optogenetics to interrogate self-organization in stem cells.
Selected by Pavithran RavindranCategories: cell biology, synthetic biology
Background
Cells are constantly bombarded with extracellular ligands and to encode this information they activate signaling pathways in distinct time-varying patterns. How such signals are interpreted in order to make cell fate decisions is largely unclear1,2. The ability to directly test how such signals are interpreted hinges on ones ability to precisely control particular signaling pathways using experimental methods. Optogenetics, the use of light responsive proteins to recruit signaling pathway effectors, has begun to play an essential role in filling this need. Work has been done to control particular pathways in cells, zebrafish and Drosophila embryos3. In this work, the authors decide to further their previously published optogenetic tool to control WNT signaling by integrating the system into stem cells4.
Key Findings
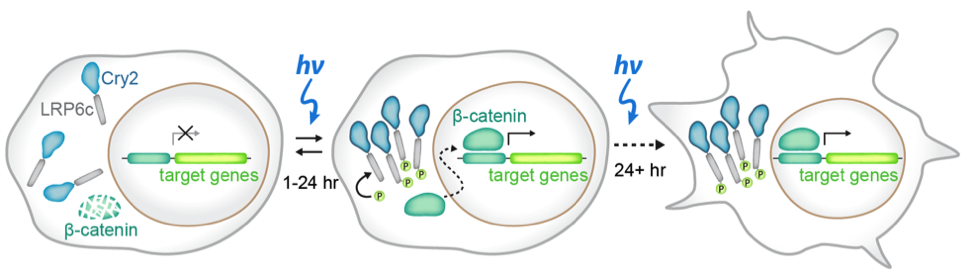
In this preprint, the authors set out to develop a method to study stem cells to study how spatial and temporal signaling dynamics regulate cell fates. Classical methods in which scientists use exogenous ligands or inhibitors to study signaling pathways gives poorly defined spatial resolution (every cell is treated the same) and temporal resolution (it is difficult to toggle a pathway on and off). To overcome this issue, the authors use a previously published optogenetic tool that specifically activates the WNT signaling pathway by using a blue-light inducible clustering system known as Cry2 attached to the WNT responsive receptor LRP6, what they called the OptoWNT system4 (figure 1). After integrating the OptoWNT system into the AAVS1 safe harbor locus of human embryonic stem cells (hESCs), the authors validate that blue light causes reversible clustering of LRP6, and accumulation of the Wnt transcriptional activator beta-catenin. From there, the authors wanted to ensure that Wnt activation through optogenetics in these stem cells recapitulates the known differentiation after recombinant Wnt3a activation. By comparing levels of Brachyury, a master mesendoderm transcription factor and marker of the primitive streak, in cells that got no light or constant 48 hours of blue light exposure, the authors found that there was a 40 fold increase in protein expression with blue light. Finally, by performing RNA-sequencing on both WT hESCs and hESCs integrated with the OptoWNT system in dark and blue light conditions, the authors found that there was very little photo-toxicity from continuous blue light exposure, low OptoWNT dark state activity and induction of differentiation upon blue light stimulation. All of these results suggest that the optogenetic tool for WNT activation integrated into the AAVS1 locus functions in a blue light dependent manner.
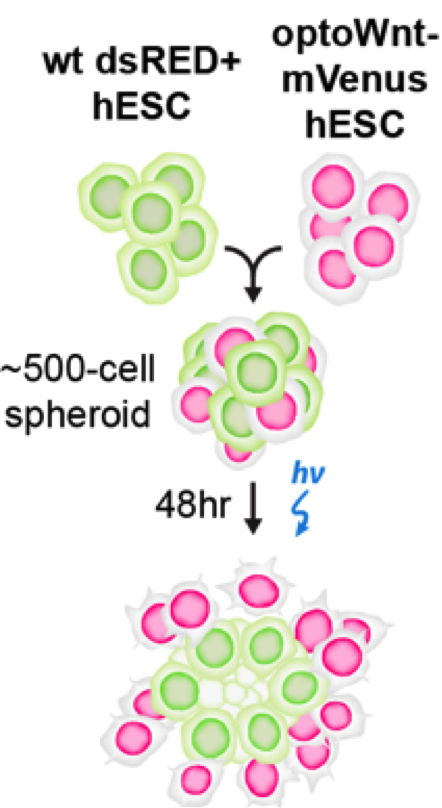
Once the authors were able to validate the OptoWNT system in hESCs, they wanted to address a question that classic drug/growth factor additions could not: what effect does the activation of WNT have when only a subset of cells in a population are “listening” for this activation? To answer this, they mixed wild-type hESCs and hESCs with OptoWNT (which were also mCherry positive such that they could distinguish between the two populations) and then applied blue light. They found that upon this blue light stimulation, the mCherry OptoWNT hESCs would segregate out and thus would have more mCherry positive neighbors as opposed to WT neighbors. They found that this segregation occurred in almost any media they checked, including media without FGF2 and TGFβ, suggesting that WNT activation alone was sufficient to drive this self-organization. Even more impressively, the authors found that self-organization occurred in a 3D co-culture of wild-type and OptoWNT hESCs in which the OptoWNT activated cells formed a ring around the wild-type cells (figure 2). All of these results suggest that WNT activation in a subset of cells may be sufficient to drive self-organization of cells.
Why I chose this preprint
Optogenetics is an amazing tool that allows researchers spatiotemporal control of signaling pathway activation in cells. Sadly however, the tool has been used mostly in either immortalized cell lines, which are far from normal biological representatives, or full organisms such as the embryo, which take a long time to develop. This work seems to hit the sweet spot directly in between these extremes by applying optogenetics to an interesting biological system that also has the advantages of normal cell culture. With this work, it will be extremely interesting to see what biologists will be able to learn about stem cells and the paths they take towards differentiation.
Questions for the authors
- In this work you have done a great job is showcasing the utility of optogenetics for the selective activation of a subset of cells within a population. Another great use of this tool is in the temporal dimension. Were you able to apply dynamic pulses of blue light to these cells and did this have any effect on the self-organization that you have described in your work?
- You have proposed a model in which WNT activation in a subset of cells is sufficient to drive self-organization, possibly for a developmental case. Do you know of any cases in which this may occur in the early embryo? Do you have a hypothesis as to the mechanism of only certain cells being receptive to the input?
References
- Purvis, J. E. & Lahav, G. Encoding and decoding cellular information through signaling dynamics. Cell 152, 945–956 (2013).
- Maryu, G. et al. Live-cell Imaging with Genetically Encoded Protein Kinase Activity Reporters. 74, 61–74 (2018).
- Johnson, H. E. & Toettcher, J. E. Illuminating developmental biology with cellular optogenetics. Curr. Opin. Biotechnol. 52, 42–48 (2018).
- Bugaj, L. J., Choksi, A. T., Mesuda, C. K., Kane, R. S. & Schaffer, D. V. Optogenetic protein clustering and signaling activation in mammalian cells. 10, 249–252 (2013).
Posted on: 30 July 2019 , updated on: 23 August 2019
doi: https://doi.org/10.1242/prelights.12654
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cell biology category:
Fetal brain response to maternal inflammation requires microglia
Alteration of long and short-term hematopoietic stem cell ratio causes myeloid-biased hematopoiesis
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Also in the synthetic biology category:
Discovery and Validation of Context-Dependent Synthetic Mammalian Promoters
Genetically encoded multimeric tags for intracellular protein localisation in cryo-EM
Dissecting aneuploidy phenotypes by constructing Sc2.0 chromosome VII and SCRaMbLEing synthetic disomic yeast
preLists in the cell biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |
Also in the synthetic biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Advances in Drug Delivery
Advances in formulation technology or targeted delivery methods that describe or develop the distribution of small molecules or large macromolecules to specific parts of the body.
| List by | Zhang-He Goh |











 (2 votes)
(2 votes)