Organoid Easytag: an efficient workflow for gene targeting in human organoids
Preprint posted on 5 May 2020 https://www.biorxiv.org/content/10.1101/2020.05.04.076067v1
Article now published in eLife at http://dx.doi.org/10.7554/eLife.67886
Gene-targeting for all: expanding the applications of CRISPR in organoid research
Selected by Kirsty FergusonCategories: developmental biology, molecular biology
Background
In recent years, CRISPR/Cas9 technology has revolutionised our ability to modify the genome of mammalian cells, in a much quicker and more reliable manner than previously. This technique adds an invaluable tool to our repertoire of genetic modification methods, expanding our ability to functionally annotate genomes using targeted genetics. While most advanced in two-dimensional cell culture systems, there is a need for robust protocols to apply this technology to more complex 3D conditions.
Organoids are self-organising 3D structures that recapitulate aspects of organ structure and function. They are increasingly used to study disease and development, as well as holding promise for translational purposes. The repair of CRISPR/Cas9-induced DNA breaks is mediated by either non-homologous end-joining (NHEJ) or, in the presence of a repair template, homology-directed repair (HDR). NHEJ-mediated repair has been used to successfully edit intestinal organoids to model colorectal cancer (1,2), as well as kidney disease (3). However, until recently, this method of repair has been associated with the error-prone introduction of insertions and deletions (4). While HDR has been used to correct a precise mutation in the CFTR gene of intestinal stem cell organoids of CF patients (5), it suffers from low efficiency compared to the NHEJ pathway and is dependent on cells being in S phase (6). Researchers are therefore searching for ways to enhance HDR or inhibit NHEJ, to facilitate precise and efficient gene modification. Here, the authors describe an HDR-mediated pipeline, for high efficiency knock-ins in human foetal lung organoids.
Key findings
Optimisation of the ‘Organoid Easytag’ pipeline
i) Optimising transfection & Cas9/gRNA/DNA delivery methods
Efficient gene targeting by HDR is dependent on multiple factors: efficient delivery of and cleavage by a Cas9/gRNA complex at the target site, efficient delivery of the donor template to provide a sufficient concentration at the time of repair, the length of donor template homology arms, the cell cycle stage, and the activity of the endogenous repair systems (7,8). To optimise the delivery of CRISPR reagents, as well as the efficiency of site-specific DNA cleavage, the authors first assessed i) different transfection methods and ii) different forms of Cas9/gRNA complex delivery. They concluded that i) nucleofection, and ii) a pre-assembled ribonucleoprotein (RNP) made up of Cas9 protein and a synthetic single-strand gRNA (ssRNP), provides the highest transfection efficiency.
Following this, the authors used ACTB, a gene encoding an abundant cell-cell junction protein in human lung organoids, to optimise their pipeline. A pre-assembled ssRNP, together with a circular plasmid repair template, were delivered by nucleofection into single lung organoid cells. Clonal organoid lines, expressing a correctly localised N-terminal EGFP-ACTB fusion protein, could be successfully generated at high efficiency following flow cytometry-based enrichment of EGFP-positive cells. Assessment of agonists and antagonists of the HR and NHEJ pathways, respectively, revealed no increase in gene targeting efficiency.
ii) Comparison of plasmid-based and ssODN repair templates
Plasmid-based repair templates enable large DNA inserts and lengthier homology arms, however require vector production. Single-stranded oligonucleotide donors (ssODNs), in contrast, provide a ‘cloning free’ workflow, but are limited in length. To investigate the use of ssODNs, the authors adopt the split GFP system, whereby part of GFP (GFP11) is provided on the ssODN or repair plasmid, and the remainder (GFP(1-10)) on a transient expression plasmid. GFP-positive cells were recorded by flow cytometry in both instances, however organoid colonies did not recover from ssODN-transfected cells. Subsequent ssODN-mediated epitope-tagging of the transcription factor SOX2 revealed that a successfully targeted heterozygous clone contained a random insertion near the gRNA target site, suggesting ssODNs lead to error-prone HDR in this system (9).
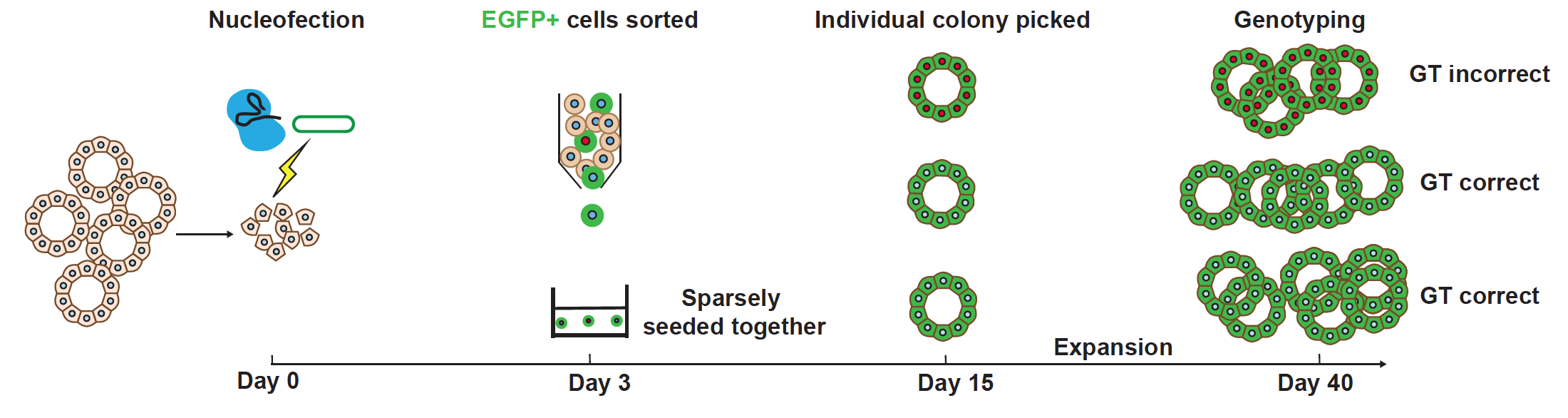
Schematic of Organoid Easytag workflow for EGFP-ACTB fusion. From Figure 1a of this preprint, made available under a CC-BY-NC-ND 4.0 International license.
Applications of the knock-in pipeline at various gene loci
i) Fluorescent-tagging of the transcription factor, SOX9
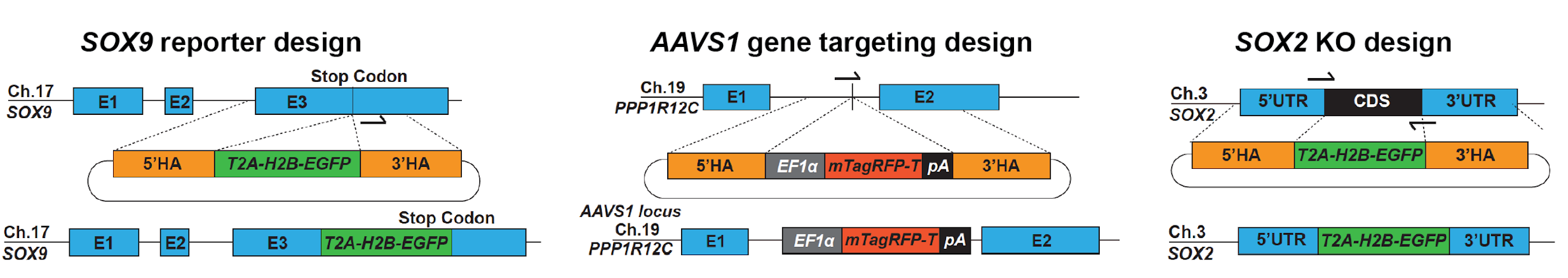
Using this optimised pipeline, the authors demonstrate the generation of heterozygous SOX9 reporter lines, a tip progenitor marker in the lungs and useful reporter of the lung progenitor state. Interestingly, the authors generate a self-cleaving SOX9-T2A-Histone2B-EGFP fusion protein to enable nuclear enrichment, overcoming the low expression of SOX9 whilst ensuring the protein is minimally influenced by the large protein tag.
ii) Knock-in at the AAVS1 human safe-harbour locus
To demonstrate the applicability of this pipeline to other genomic loci, a membrane-tagged, monomeric red fluorescent protein sequence (TagRFP-T) was integrated under the control of an EF1α promoter at the AAVS1 locus. This provides a high efficiency method to knock-in and control the expression of exogenous genes at a human safe-harbour locus.
iii) Knock-out of the transcription factor, SOX2
The generation of gene-knockouts can be complicated by i) in-frame exon skipping creating alternative isoforms, and ii) difficulties in enrichment for the knock-out population. When transfection efficiencies are low and a selectable marker is absent, transfected cells must be screened at a clonal level by PCR genotyping or protein detection. Here, the authors sequentially knock-out both copies of SOX2 by replacing the coding sequence with the T2A-H2B-EGFP reporter sequence, enabling the identification and isolation of EGFP-positive SOX2-/- cells by flow cytometry.
Schematics showing repair template design and final products for SOX9 reporter line, AAVS1 targeting and SOX2 KO. Adapted from Figure 2 of this preprint, made available under a CC-BY-NC-ND 4.0 International license.
Why I chose this preprint and what I liked about it
Organoids have an increasingly important role in modelling the development and diseases of human tissues in vitro, as well as exciting potential roles in regenerative medicine. There is therefore a need to develop robust systems to study and modify genes in these 3D systems. While CRISPR has been used to investigate gene function in lung organoids previously (10), this pipeline expands the genetic modifications that can be achieved at high efficiency in this system. As all plasmids used in this study will be deposited in Addgene, this provides new tools for the field and opens up new research avenues.
It is also interesting how this study provides a complementary gene knock-in approach to the recently published NHEJ-mediated CRISPR-HOT method (11), with both studies circumventing the need for TP53 inhibition to increase HDR efficiency. I enjoyed reading about their approaches to use of an H2B-EGFP fusion to overcome the low expression of SOX9 and the replacement of SOX2 with T2A-H2B-EGFP enabling enrichment of successfully targeted clones without lengthy screening protocols.
In my research I perform CRISPR/Cas9-mediated gene knock-outs and gene-tagging in 2D neural stem cell and brain cancer stem cell cultures. Similarly, improved efficiencies of CRISPR editing using recombinant Cas9 protein have been seen in this system (12,13). In contrast, two-part cr/trRNA have been found to show increased transfection efficiencies compared to sgRNA for epitope knock-in (13). I was interested to see how these methods differ in an organoid system.
Questions for the authors
- Do you ever obtain mosaic organoids containing both correctly and incorrectly-targeted cells? As genotyping would reveal wild-type and targeted alleles in the case of both knock-in heterozygosity and mosaicism, does immunostaining of organoids allow you to assess a sufficient number of cells to distinguish these scenarios?
- In Figure 2e, is the targeting efficiency collated for all organoid lines tested? If so, is this efficiency roughly equal between lines or does it vary?
- Compared to the two-part cr/trRNA system, synthetic sgRNAs are less adaptable and more costly (13). If you were to increase the throughput of the Organoid Easytag pipeline, would you consider the increase in efficiency that sgRNAs offer to justify their use?
- Were both ssRNA (Synthego) and cr/trRNAs (IDT) chemically modified to limit cellular immune responses and increase stability? (12) Have you found incubating the cr/trRNA at 95°C for 2 minutes sufficient (compared to 5 minutes recommended by IDT)?
- When describing the number of organoid lines tested per experiment, were each of these lines tested as independent experiments to account for technical variations?
- Did you consider comparing the use of linear dsDNA alongside ssODNs and plasmid donors for knock-in of larger inserts? (14,15)
- In supplementary figure 3, did you consider using homology arms of the same length in both the ssODN and plasmid donor? Would this help to distinguish if the difference in transfection efficiency is a result of linear single-stranded DNA versus circularised dsDNA, or rather the homology arm length? It seems that fewer ‘ssODN-transfected’ than ‘plasmid-transfected’ cells were recovered following flow sorting – were the same number of cells plated to ensure the difference in organoid formation is a product of the ssODN treatment, rather than poor recovery at a low plating density? Is it possible that a ssODN concentration of 500 pmol is toxic for the cells – did you consider testing a lower concentration of ssODN? (15)
- Related to this, would it be possible to recover, and perform indel analysis, on the GFP-positive ssODN-transfected cells following sorting, to ascertain if tagging was also error-prone in this experiment? Would it be beneficial to isolate additional SOX2-V5 clones to confirm that indel formation is a recurring event when using ssODN donor DNA in this system?
- Would it be possible to devise a strategy for efficient generation of biallelic knockouts with a single round of transfection and selection? For example, could one SOX2 CDS be replaced by the T2A-H2B-EGFP reporter and the non-targeted allele be disrupted by more efficient NHEJ-mediated indel formation (strategy used in 13)? In fact, is it possible NHEJ-mediated disruption of the non-targeted allele could have occurred prior to re-targeting?
- Did you test the efficiency when transfecting whole organoids, rather than single cells? As well as founder cells, do you envision this pipeline being applied to established organoids (e.g. more complex cerebral organoids) without cell dissociation?
- How do you plan to implement this technology next in your research?
Thank you to the authors for responding to these questions – see their thoughtful answers below.
References
(1) Drost J, van Boxtel R, Blokzijl F, Mizutani T, Sasaki N, Sasselli V, de Ligt J, Behjati S, Grolleman JE, van Wezel T, et al (2017) Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science 358(6360):234-238
(2) Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, Sachs N, Overmeer RM, Offerhaus GJ, Begthel H et al (2015) Sequential cancer mutations in cultured human intestinal stem cells. Nature 521(7550):43-7
(3) Freedman BS, Brooks CR, Lam AQ, Fu HX, Morizane R, Agrawal V, Saad AF, Li MK, Hughes MR, Vander Werff R et al (2015) Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat Commun 6: 8715
(4) Nie J & Hashino E (2017) Organoid technologies meet genome engineering. EMBO Rep 18(3): 367–376.
(5) Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent CK et al. (2013) Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 13, 653–658
(6) Hustedt N & Durocher D (2016) The control of DNA repair by the cell cycle. Nat Cell Biol 19(1):1-9
(7) Elliott B, Richardson C, Winderbaum J, Nickoloff JA, Jasin M (1998) Gene conversion tracts from double-strand break repair in mammalian cells. Mol Cell Biol 18(1): 93-101
(8) Liu M, Rehman S, Tang X, K Gu, Fan Q, Chen D and Ma W (2018) Methodologies for Improving HDR Efficiency. Front Genet 9: 691.
(9) Boel A, De Saffel H, Steyaert W, Callewaert B, De Paepe A, Coucke PJ and Willaert A (2018) CRISPR/Cas9-mediated homology-directed repair by ssODNs in zebrafish induces complex mutational patterns resulting from genomic integration of repair-template fragments. Dis Model Mech 11(10): dmm035352.
(10) Gao X, Bali AS, Randell SH and Hogan BLM (2015) GRHL2 coordinates regeneration of a polarized mucociliary epithelium from basal stem cells. J Cell Biol 211(3): 669–682.
(11) Artegiani B, Hendriks D, Beumer J, Kok R, Zheng X, Joore I, de Sousa Loupes SMC, van Zon J, Tans S and Clevers H (2020) Fast and efficient generation of knock-in human organoids using homology-independent CRISPR–Cas9 precision genome editing. Nat Cell Biol 22, 321–331
(12) Kelley ML, Strezoska Ž, He K, Vermeulen A & Smith Av (2016) Versatility of chemically synthesized guide RNAs for CRISPR-Cas9 genome editing. J Biotechnol 233:74-83.
(13) Bressan RB, Dewari PS, Kalantzaki M, Gangoso E, Matjusaitis M, Garcia-Diaz C, Blin C, Grant V, Bulstrode H et al (2017) Efficient CRISPR/Cas9-assisted gene targeting enables rapid and precise genetic manipulation of mammalian neural stem cells. Development 144: 635–648
(14) Dewari PS, Southgate B, Mccarten K, Monogarov G, O’Duibhir E, Quinn N, Tyrer A, Leitner M-C, Plumb C, Kalantzaki M et al (2018) An efficient and scalable pipeline for epitope tagging in mammalian stem cells using Cas9 ribonucleoprotein. Elife 7: 1–29
(15) Li H, Beckman KA, Pessino V, Huang B, Weissman JS & Leonetti MD. Design and specificity of long ssDNA donors for CRISPR-based knock-in. bioRxiv 178905: doi: https://doi.org/10.1101/178905
Posted on: 22 May 2020
doi: https://doi.org/10.1242/prelights.20966
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the developmental biology category:
Temporal constraints on enhancer usage shape the regulation of limb gene transcription
OGT prevents DNA demethylation and suppresses the expression of transposable elements in heterochromatin by restraining TET activity genome-wide
Actin polymerization drives lumen formation in a human epiblast model
Also in the molecular biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Nanos2+ cells give rise to germline and somatic lineages in the sea anemone Nematostella vectensis
Plant plasmodesmata bridges form through ER-driven incomplete cytokinesis
AND
Plasmodesmata act as unconventional membrane contact sites regulating inter-cellular molecular exchange in plants
preLists in the developmental biology category:
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the molecular biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (No Ratings Yet)
(No Ratings Yet)