Plant photoreceptors and their signaling components compete for binding to the ubiquitin ligase COP1 using their VP-peptide motifs
Preprint posted on 5 March 2019 https://www.biorxiv.org/content/10.1101/568618v1
Article now published in The EMBO Journal at http://dx.doi.org/10.15252/embj.2019102140
Shedding light on photoreceptor action: plant photoreceptors interfere with substrate binding of the light signalling repressor COP1 by direct competition for binding sites
Selected by Martin BalcerowiczCategories: biochemistry, plant biology
Background: Plant light signalling converges on the repressor COP1
Plants perceive light with a diverse set of photoreceptors, among them the red/far-red light-sensing phytochromes (phyA-E), the blue light-sensing cryptochromes (cry1, cry2) and the UV-B-sensing UV RESISTANCE LOCUS 8 (UVR8). The signalling cascades downstream of these photoreceptors converge on the regulation of a central repressor of light signalling, a complex of CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) and SUPPRESSOR OF PHYA-105 (SPA) proteins. In darkness, this COP1/SPA complex ubiquitinates positive regulators of the light response and thereby targets them for degradation via the 26S proteasome. In the light, photoreceptors inhibit COP1/SPA activity, allowing positive regulators of the light response to accumulate and promote light-induced gene expression1.
Several mechanisms have so far been described for photoreceptors to control COP1/SPA activity1,2: (1) Prolonged exposure to light leads to exclusion of COP1 from the nucleus, which is dependent on phyA, phyB and cry1; (2) phyA and phyB promote proteasomal degradation of SPA proteins upon light exposure; (3) light-activated phyA, phyB and cry1 reduce interaction between COP1 and SPAs, thereby disrupting the COP1/SPA complex. UVR8 and cry2 are also known to interfere with COP1/SPA activity through direct interaction, but the underlying processes remain unclear. In this preprint, Lau, Podolec et al. show that these two photoreceptors directly interfere with the binding of COP1 to its target proteins.
Key findings: Light-activated photoreceptors compete with downstream transcription factors for binding to COP1
A so-called VP motif (containing Val and Pro as key residues), first described in the transcription factor ELONGATED HYPOCOTYL 5 (HY5)3, is known to mediate interactions with the COP1 WD40 domain. HY5 is a downstream component in UV-B signalling and is stabilised upon UV-B-induced binding of the UVR8 photoreceptor to COP1.4 Intriguingly, UVR8 contains a VP motif in its C-terminus, a domain essential for UV-B signalling. By employing in vitro binding assays and crystal structure analyses the authors found that both UVR8 and HY5 VP-peptides bind to the same site in the COP1 WD40 domain; the Val and Pro residues establish hydrophobic interactions to the same aromatic residues in COP1 while an upstream Tyr (UVR8) or Arg (HY5) residue form hydrogen bonds and salt bridges to COP1 polar residues (Fig. 1). Mutating these core residues in the COP1 protein abolished or greatly reduced binding to UVR8 and HY5 and, in agreement with this, the respective COP1 mutant variants could not restore UV-B signalling in a cop1 mutant background.
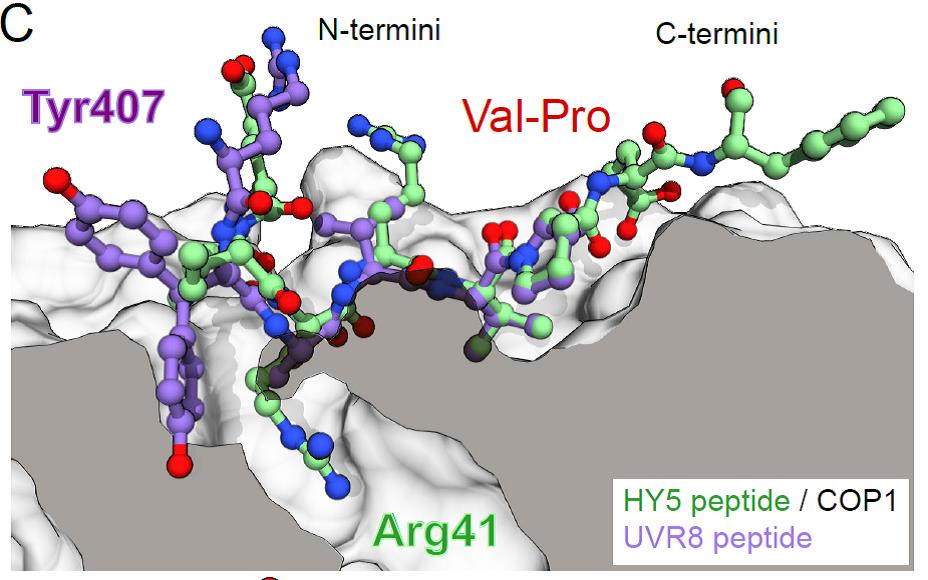
Figure 1: Superposition of the X-ray crystal structures of HY5 and UVR8 peptides in the VP-peptide binding site of the COP1 WD40 domain. COP1 is depicted in surface representation, HY5 and UVR8 peptides are depicted in ball-and-stick representation in green and purple, respectively. (reproduced from Lau, Podolec et al., Fig. 1 C, under a CC-BY-NC-ND 4.0 license).
UVR8 forms homodimers, but monomerises upon UV-B irradiation and binds to COP1 to promote UV-B responses4. The fact that UVR8 and HY5 VP-peptides occupy the same binding site in COP1 implies that they may compete for binding, representing a possible mechanism for UVR8 to repress COP1 activity. Binding of the HY5 peptide to COP1 was approximately 8 times stronger compared to the UVR8 peptide in vitro, arguing against effective displacement of HY5 by the photoreceptor. However, performing binding assays with full-length proteins revealed that UV-B irradiation strongly enhances binding of UVR8 to COP1 due to cooperative binding of both its VP region and its beta-propeller core to the COP1 WD40 domain. Binding strength of full-length HY5 to COP1 was in a similar range to that of non-photoactivated UVR8, suggesting that only photoactivated UVR8 can efficiently displace HY5 at the COP1 VP binding site. Yeast-3-hybrid assays further confirmed that COP1 interacts with HY5 in the absence of UV-B, but that UV-B irradiation abolished their interaction when UVR8 is present.
The VP mode of interaction appears to be conserved in other COP1 interaction partners. The authors tested a set of transcription factors such as the floral promoter CONSTANS (CO) as well as the photoreceptors CRY1 and CRY2, all of which harbour a VP motif. Crystal structures revealed that the respective VP peptides all bind to the VP binding site in COP1 in a similar configuration – the Val and Pro residues occupy the centre of the binding site, while chemically diverse residues 2-3 positions upstream help anchor the peptide. The CRY2 and CO interactions are particularly interesting as CRY2 is known to promote flowering under long day conditions by stabilising the CO protein through repression of COP12. Binding of full-length CRY2 to COP1 was strongly enhanced upon photoactivation, suggesting that CRY2 may act in a similar way to UVR8, displacing CO and preventing its degradation.
What I like about this preprint
This preprint represents a great example of how multidisciplinary approaches, exemplified here by crystallographic, biochemical and genetic characterisations, can bring about a leap in understanding of molecular signalling processes. This is especially noteworthy in context of photoreceptors and the COP1/SPA complex, whose relations have been studied for decades, yet are still not fully understood. Lau, Podolec et al. brought us a significant step closer to decipher the mechanisms underlying this relationship.
Open questions/future directions
- Earlier studies described a positive role for COP1 in the UV-B signalling pathway, evidenced by the absence of certain UV-B responses in a cop1 mutant5. However, more recent studies, including the results of Lau, Podolec et al., consolidate the role of COP1 as a negative regulator of light signalling downstream of phytochromes, cryptochromes and UVR8. In light of these findings, do we still think that COP1 has a unique (possibly dual) role downstream of UVR8, or can we consider its function to be broadly the same in the UV-B and visible light signalling pathways?
- The COP1 VP binding site appears to be highly plastic and thus able to accommodate variable VP peptides. Is it possible that other photoreceptors besides UVR8 and cryptochromes bind to it and thereby affect COP1 activity in the same way?
References/Further reading
- Hoecker U (2017). The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling. Curr Opin Plant Biol. 37:63-69.
- Podolec R, Ulm R (2018). Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr Opin Plant Biol. 45:18–25.
- Holm M, Hardtke CS, Gaudet R, Deng XW (2001). Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 20: 118–127.
- Liang T, Yang Y, Liu H (2019). Signal transduction mediated by the plant UV-B photoreceptor UVR8. New Phytologist 221: 1247–1252
- Oravecz A, Baumann A, Máté Z, Brzezinska A, Molinier J, Oakeley EJ, Adám E, Schäfer E, Nagy F, Ulm R. (2006). CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell. 18:1975-90.
Posted on: 25 March 2019
doi: https://doi.org/10.1242/prelights.9613
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biochemistry category:
Structural basis of respiratory complexes adaptation to cold temperatures
Lens Placode Modulates Extracellular Matrix Formation During Early Eye Development
Generalized Biomolecular Modeling and Design with RoseTTAFold All-Atom
Also in the plant biology category:
Plant plasmodesmata bridges form through ER-driven incomplete cytokinesis
AND
Plasmodesmata act as unconventional membrane contact sites regulating inter-cellular molecular exchange in plants
H2O2 sulfenylates CHE linking local infection to establishment of systemic acquired resistance
Coordinated regulation of photosynthesis and translation via NIK1/RPL10/LIMYB signaling module in response to biotic and abiotic stresses
preLists in the biochemistry category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
Preprint Peer Review – Biochemistry Course at UFRJ, Brazil
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biochemistry deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the plant biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |











 (No Ratings Yet)
(No Ratings Yet)