Precise genomic deletions using paired prime editing
Preprint posted on 2 January 2021 https://www.biorxiv.org/content/10.1101/2020.12.30.424891v1
Article now published in Nature Biotechnology at http://dx.doi.org/10.1038/s41587-021-01025-z
PRIME-Del: Two “pegs” are better than one – precise genome deletions with paired prime editing guide RNA
Selected by Fabio LiberanteCategories: genomics, molecular biology, synthetic biology
Context
Genome engineering has seen an explosion in recent years thanks to the Nobel prize-winning discovery of CRISPR/Cas proteins. Their ease of use has enabled a myriad of new functional studies exploring genes, domains, enhancers and structural elements of DNA sequence. The original programmable DNA-binding technology has spawned numerous novel molecular tools, including activators, inhibitors, silencers, repressors, interaction-creators and base editors. At the end of 2019 a new CRISPR tool took centre-stage, prime editing. This ingenious use of a Cas-reverse transcriptase fusion and an extended guide RNA (pegRNA) enabled very simple substitutions, insertions and deletions at defined loci, no longer relying on DNA donors or limited by the one-at-a-time transitions or transversions of base editors. Another advantage is the fact that the site to be targeted and the template are encoded in a single pegRNA. The paper has already received over 200 citations in just over 12 months showing its usefulness. Despite its potential, it’s still unable to delete large regions efficiently. Similarly, the use of paired guides with typical CRISPR/Cas to trigger simultaneous double-strand breaks is fraught with its own problems. The authors of this preprint present an application of prime editing that enables large precise deletions and simultaneous insertions along with a tool for designing the necessary pegRNAs.
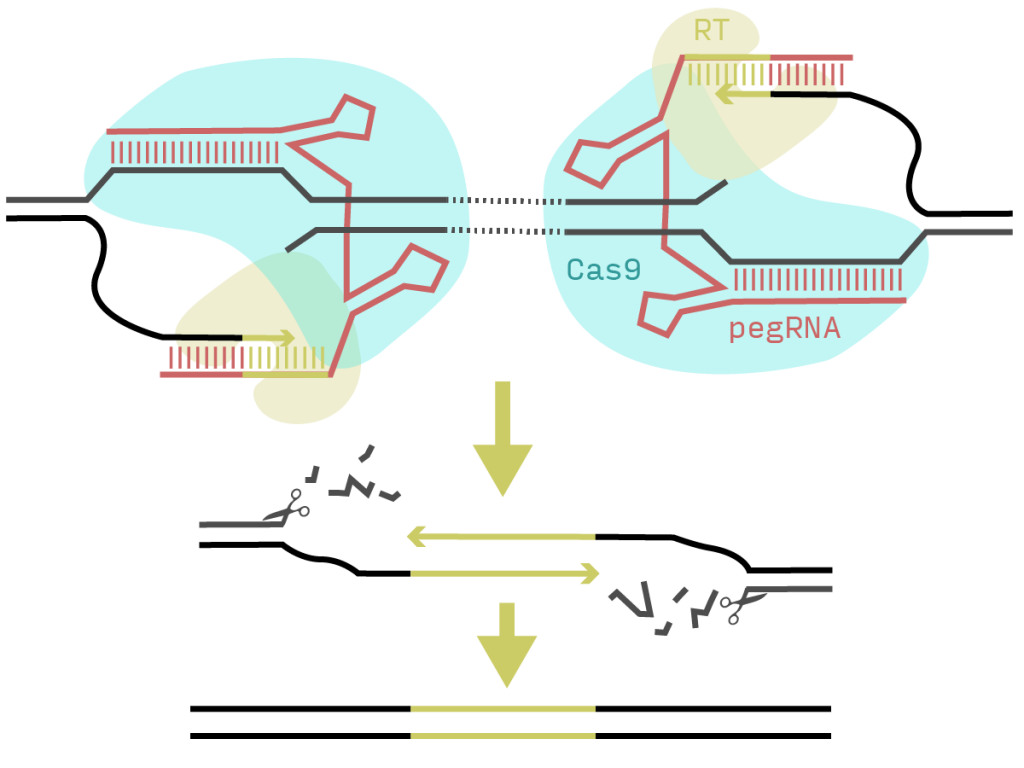
Key ideas
PRIME-Del – creating deletions with prime editing and pairs of pegRNAs
Prime editing uses a Cas9 nickase and reverse transcriptase (RT) fusion (PE2) together with an extended guide RNA (pegRNA). The Cas9 nicks the DNA and the RT extends the free end using the 3’ region of the pegRNA as primer and template. The authors of this preprint realised that they could use a second pegRNA on the opposite strand at an arbitrary distance and insert a matching overhang that should trigger a base-pair-accurate deletion of the intervening region (Figure 1).
To test this approach they assembled a dual pegRNA vector using the U6 and H1 RNA PolIII promoters on the same plasmid. Initially, they targeted the eGFP cassette present on the prime editor (PE2) expressing vector. The PRIME-Del method proved successful and showed an efficiency ranging between 38% and 77% for 24 to 546bp deletions for this episomal target. Interestingly, larger deletions showed a higher efficiency.
Once the authors had shown the system worked they attempted to use it to modify a genomic target and so integrated a similar eGFP cassette using lentivirus. While successful, this was unfortunately much less efficient at only 10%. However, targeting the HPRT1 native genomic locus yielded much better deletion rates of up to 25%. This likely illustrates the importance of the target and its genomic context.
PRIME-Del is more precise than CRISPR/Cas9
The authors next compared PRIME-Del to previous techniques, which use a pair of regular guide RNAs and double-strand cleaving Cas9. This showed that Cas9 yielded ~15% indels even without successful deletion of the target region vs only 2.2% for PRIME-Del. The overall error rate for both was higher if the region was deleted, but still nearly four times lower for PRIME-Del. While the much greater precision of the PRIME-Del approach is impressive, most use cases for cells modified in such a way requires sub-cloning to ensure that only cells with the exact mutation/deletion are studied. When the efficiencies for both systems are looked at purely in terms of the overall percentage of cells with the exact deletion they are not dissimilar. In the example presented for the 252 bp deletion in HPRT1 the percentage of perfect deletions are approximately 25% vs. 19% for PRIME-Del and Cas9, respectively. So while PRIME-Del is very precise, it is only marginally more efficient than Cas9, despite its more complex design.
I suspect the authors were disappointed with the small increase in efficiency initially offered by PRIME-Del. Therefore they attempted to boost it by constitutive expression of both the prime editor and the dual pegRNAs. This is made viable by the high precision of the system as random “dead-end” mutations don’t accumulate quickly. Promisingly, this proved a success and yielded correct 118 bp deletions in nearly 50% of K-562 cells after 19 days in culture.
Benefits of PRIME-Del
Despite the limited overall efficiency in transient PRIME-Del, there are certain circumstances where greater precision is indeed much more important. One obvious example is in any future applications in gene therapy – where any unintended mutations could be dangerous. While the authors do not directly recommend PRIME-Del for use in gene therapy, they demonstrate its potential by deleting a CGG-repeat expansion region in the FMR1 gene implicated in Fragile-X syndrome. A second example where precision is important is in any multiplex studies where single cell cloning is not possible – for example if targeting various protein domains for deletion, reducing the number of spurious indels or knockouts simplifies downstream analysis. Further, PRIME-Del, and prime editing in general, are likely better suited to high-throughput screening as they only induce single-strand DNA nicks, which is often less toxic to cells, a common limiting factor to using regular CRISPR/Cas in certain cell types.
Perhaps one of the most exciting innovations of the PRIME-Del system is that it allows simultaneous insertion of novel sequence at the point of deletion. The authors demonstrated successful insertion of up 30bp using this technique. It doesn’t rely on a separate repair template and does not require homologous repair to incorporate foreign DNA. Encouragingly, the authors show that if the deletion is successful then so is the insertion in over 99% of cases.
Challenges and limitations
In terms of pitfalls, the authors identify some designs whereby PRIME-Del can trigger a relatively high frequency of small duplications, mainly due to high GC content at the 3’ ends of the engineered overlaps. This is something that their design tool seeks to minimise.
While the design of a pegRNA is more complicated than a regular guide RNA and a paired pegRNA PRIME-Del design even more so, the authors have helpfully written a Python-based tool [https://github.com/shendurelab/Prime-del] (and web instance https://primedel.uc.r.appspot.com/]) which simplifies the process.
The authors do highlight some limitations with their system. Primarily, that paired pegRNAs require the presence of PAM sites near each end of the deletion and on opposing strands. They suggest that this might eventually be solved using a “PAM-less” prime editor.
What I like about this work
One of the great things about this work is the simplicity of the idea, which is then borne out in a series of well thought-out experiments. Authors addressed a typical concern about PCR bias in quantifying deletion efficiency by using a clever linear amplification step to label molecules with a 15 bp UMI – it turned out there were only modest effects anyway! PRIME-Del is tested in the proper range of contexts – episomal, integrated synthetic construct and native genomic loci. Finally, the preprint comes with an accompanying software tool to enable others to quickly design and test their own PRIME-Del pegRNAs.
Open questions
- The largest deletion demonstrated was 710 bp. What are the upper size limits for PRIME-Del?
- Simultaneous insertion at the point of deletion is useful, but what is the largest insert?
- The authors mention that PAM sites need to be present near deletion junctions, but how far away can the sites be? Especially important in AT-rich genomes.
- Did the authors consider trying a multiple transfection (or sustained expression) approach with regular Cas9 to demonstrate the “dead ends” that should accumulate? Also, did they compare PRIME-Del to a single pegRNA for deletions?
- The authors used H1 and U6 promoters to express the two pegRNAs. Did they try other PolIII promoters, such as mU6 or 7SK, or even Csy4 arrays?
- Interestingly, it seems that in most cases larger deletions were more efficient. Do the authors have any suspicions as to why? Is it simple steric hindrance?
- The overall efficiency seems to vary widely depending on cell type and genomic context. Did the authors investigate what factors might contribute to this variation? Guide RNA score, chromatin structure etc.?
Posted on: 27 January 2021
doi: https://doi.org/10.1242/prelights.27128
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the genomics category:
Temporal constraints on enhancer usage shape the regulation of limb gene transcription
Transcriptional profiling of human brain cortex identifies novel lncRNA-mediated networks dysregulated in amyotrophic lateral sclerosis
A long non-coding RNA at the cortex locus controls adaptive colouration in butterflies
AND
The ivory lncRNA regulates seasonal color patterns in buckeye butterflies
AND
A micro-RNA drives a 100-million-year adaptive evolution of melanic patterns in butterflies and moths
Also in the molecular biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Nanos2+ cells give rise to germline and somatic lineages in the sea anemone Nematostella vectensis
Plant plasmodesmata bridges form through ER-driven incomplete cytokinesis
AND
Plasmodesmata act as unconventional membrane contact sites regulating inter-cellular molecular exchange in plants
Also in the synthetic biology category:
Discovery and Validation of Context-Dependent Synthetic Mammalian Promoters
Genetically encoded multimeric tags for intracellular protein localisation in cryo-EM
Dissecting aneuploidy phenotypes by constructing Sc2.0 chromosome VII and SCRaMbLEing synthetic disomic yeast
preLists in the genomics category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the molecular biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the synthetic biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Advances in Drug Delivery
Advances in formulation technology or targeted delivery methods that describe or develop the distribution of small molecules or large macromolecules to specific parts of the body.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)