Refolding activity of bacterial Hsp90 in vivo reveals ancient chaperoning function
Preprint posted on 5 November 2018 https://www.biorxiv.org/content/early/2018/11/05/462549
Why the need for Hsp90? Clever experiments reveal the importance of this chaperone in refolding proteins after heat shock in E. coli.
Selected by Tessa SinnigeCategories: biochemistry
Background
Protein folding inside the living cell is assisted by molecular chaperones. Many of these are called heat shock proteins, since they are not only involved in aiding protein folding after synthesis, but are also upregulated upon heat stress in order to refold denatured proteins, and prevent misfolding and aggregation (1). Whereas the function and mechanisms of heat shock proteins such as Hsp70 have been well characterised, the function of Hsp90 is less clear. Hsp90 is highly abundant both in prokaryotes and eukaryotes, signifying its conserved and critical cellular role. Previously, the Rüdiger lab showed that Hsp90 is required to alleviate the folding block caused by Hsp70, which actually can prevent refolding by binding to and occluding critical hydrophobic sites in the substrate (2). In the current preprint, the authors address the question under which cellular circumstances bacterial Hsp90 is required for efficient folding.
Results
To assess protein folding in E. coli, the authors established an elegant assay based on the recombinant expression of luciferase, which produces light only when correctly folded. Hsp90 inhibitors were added either before luciferase expression was induced, or at the onset of a heat shock that left most of the luciferase unfolded (Figure A). Surprisingly, inhibiting Hsp90 just before the synthesis of luciferase was initiated did not have a large effect, showing that the role of this chaperone in the de novo folding is limited. Conversely, adding the inhibitors after a period of luciferase expression and just before the heat shock had a more dramatic effect, strongly decreasing the fraction of active luciferase in a dose-dependent manner (Figure B). This effect could be extended from luciferase to the native E. coli proteome, given that Hsp90 inhibition just before heat shock led to a larger proportion of endogenous proteins that misfolded and aggregated, as judged from SDS-PAGE band intensities of the supernatant and pellet fractions.
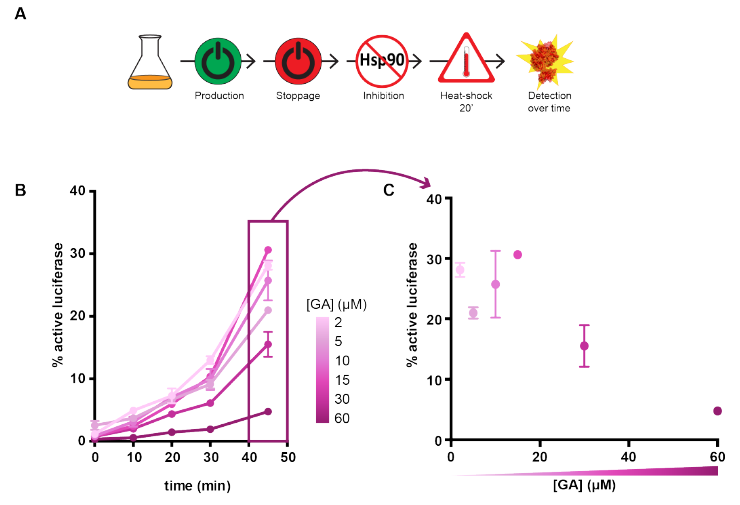
A) Design of the assay. B) Effect of varying concentrations of the Hsp90 inhibitor geldanamycin on luciferase refolding after heat shock. Reproduced from figure 2AB of the preprint under a CC-BY-NC-ND 4.0 International license.
The authors furthermore suggest that Hsp90 may stabilise the folded state of proteins, based on the observation that inhibition of Hsp90 before the start of luciferase synthesis resulted in less recovery after a brief heat shock. Finally, they hypothesised that Hsp90 may promote protein folding in the cell by reducing the overall energy consumption which would be especially important under conditions of cellular stress, and indeed they were able to show by mass spectrometry that less ATP is consumed overall if Hsp90 is added to the Hsp70 system in a reconstituted assay for luciferase refolding.
Why I chose this preprint
I really like the elegant experimental design and clear-cut results presented in this study, leading to concrete advances in our understanding of the function of Hsp90. The authors took a similar approach in their previous paper (2) and it is great to see that they were able to extend this to the in vivo environment provided by the humble model organism E. coli. In these times of continuous technological advances, big data and what not, this type of science is refreshing and I would like to see more of it!
Questions
The authors convincingly show that the global E. coli proteome depends on Hsp90 for efficient refolding after heat shock, and that the effect of Hsp90 on initial luciferase folding is rather modest. However, could it be that a subset of E. coli proteins may depend on Hsp90 for de novo folding, as has been shown in eukaryotes (1)?
Related to this, I can imagine that the ‘unfolded proteome’ of newly synthesized proteins may differ from that of denatured proteins after heat shock, which will be the less thermodynamically stable proteins. Could the authors speculate if this may influence Hsp90 dependence, i.e. if the set of heat-denatured proteins may contain more Hsp90 clients?
With respect to the authors’ previous findings, would it be possible that higher Hsp70 levels induced by the heat shock increase the demand for Hsp90 to alleviate the folding block?
I would also like to ask about the suggestion that Hsp90 stabilises the native state, based on the increased sensitivity after a brief heat shock to Hsp90 inhibition from the start of the assay. Assuming that chaperones do not change the native state of a protein compared to spontaneous folding, I would think the thermodynamic stability of folded luciferase itself would remain the same whether or not Hsp90 is inhibited. Could it just be an additive effect of the slight influence on de novo folding (which still went down to 75% at high inhibitor concentrations) combined with the reduced refolding after heat shock? Or were the data (in figure 4 of the preprint) normalised to the luciferase activity before the heat shock for each of the inhibitor concentrations?
Finally, more generally, could the authors comment on which of their findings they expect to apply to eukaryotes as well, and some of the potential differences?
References
- Kim, Y. E., Hipp, M. S., Bracher, A., Hayer-Hartl, M., and Ulrich Hartl, F. (2013) Molecular Chaperone Functions in Protein Folding and Proteostasis. Annu. Rev. Biochem.82, 323–355.
- Morán Luengo, T., Kityk, R., Mayer, M. P., and Rüdiger, S. G. D. (2018) Hsp90 Breaks the Deadlock of the Hsp70 Chaperone System. Mol. Cell70, 545–552.
Posted on: 22 November 2018
doi: https://doi.org/10.1242/prelights.5776
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biochemistry category:
Structural basis of respiratory complexes adaptation to cold temperatures
Lens Placode Modulates Extracellular Matrix Formation During Early Eye Development
Generalized Biomolecular Modeling and Design with RoseTTAFold All-Atom
preLists in the biochemistry category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
Preprint Peer Review – Biochemistry Course at UFRJ, Brazil
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biochemistry deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (No Ratings Yet)
(No Ratings Yet)