Regulatory context drives conservation of glycine riboswitch aptamers
Preprint posted on 12 September 2019 https://www.biorxiv.org/content/10.1101/766626v1
Article now published in PLOS Computational Biology at http://dx.doi.org/10.1371/journal.pcbi.1007564
How did the glycine riboswitch evolve? And where did the ghosts come from? Crum et al. show that genetic context matters in the conservation of individual parts of this riboswitch.
Selected by Defne SurujonCategories: bioinformatics, evolutionary biology
Background
Noncoding RNAs (ncRNAs) are a large group of molecules with various roles in an organism, including serving as structural components of the ribosome, tRNAs in translation and riboswitches. Riboswitches have secondary structures that allow them to respond to environmental/chemical cues and regulate gene expression. One class of riboswitches respond to glycine, and can regulate either glycine cleavage (GCV) or transport (TP). The glycine riboswitch can appear as either a single aptamer (also called singletons, which is more conventional for riboswitches) or as two aptamers in tandem. In the tandem configuration, both aptamers can bind glycine.
Previous studies have evaluated which aptamer is more functionally relevant in the tandem glycine riboswitch, and came to different conclusions. One study focused on the glycine riboswitch found in Vibrio cholerae, which is a TP-regulator, and found that the second aptamer was more relevant in controlling gene expression (Ruff and Strobel, 2014). Another study in Bacillus subtilis concluded that the first aptamer played a more important role in gene regulation; but in this case the glycine riboswitch was regulating GCV genes (Babina et al., 2017). Taken together, these seemingly contradictory results can possibly be explained by the following hypothesis: which aptamer is relevant for controlling gene expression depends on context – what organism the glycine riboswitch is found in, and what type of gene it regulates (TP or GCV). This study addresses this hypothesis by evaluating the conservation (which can be a proxy for functional relevance) of individual aptamers in different organisms, regulating different genes.
Key Findings
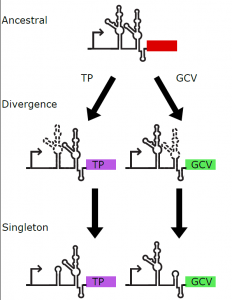
The authors first identified all glycine riboswitches, singleton and tandem, and categorized them based on their genetic context, i.e. what gene appears downstream of (and therefore might be regulated by) the riboswitch. Since existing in vitro and in vivo work had characterized V. cholerae and B. subtilis glycine riboswitch function, this study starts with a focus on the Vibrionaceae and Bacillaceae families. The glycine riboswitch does indeed appear in different contexts in the two families; upstream of genes coding for transport proteins (TP) in Vibrionaceae and of glycine cleavage system genes (GCV) for the most part in Bacillaceae. The few TP-regulating glycine riboswitches in Bacillaceae appeared to be phylogenetically more similar to the Vibrionaceae riboswitches, hinting to the importance of genetic context in shaping riboswitch evolution.
Complementing phylogenetic analysis, the authors use network clustering to evaluate the relative degree of conservation within each of the aptamers of the tandem glycine riboswitch. In this approach, riboswitch sequences are considered as vertices of a network, and each possible pair of sequences are connected with an edge based on their similarity (the authors primarily use RNAmountAlign (Bayegan and Clote, 2018) as the main similarity metric, as it takes into account both primary sequence and secondary structure). An edge connects two sequences if they are more similar than some threshold. Since the architecture of the resulting network (and therefore the conclusions we may draw) will be affected by the choice of the threshold, the authors slide the threshold from permissive (resulting in a densely connected network) to stringent (resulting in a sparse network), and consider the ensemble of these networks.
The authors split the individual aptamers of tandem riboswitches into their respective genetic contexts (TP in Vibrionaceae and GCV in Bacillaceae) and construct similarity-based networks at varying thresholds. The first and the second aptamers appear more densely connected (hence more strongly conserved) in the GCV and the TP context respectively.
What about the singletons though? Single aptamers also appear together with a “ghost” aptamer, that is potentially a degraded version of the other aptamer in the tandem that lost its secondary structure over the course of evolution. Moreover, singletons have been categorized as type 1 or type 2 based on which side of the singleton the ghost aptamer appears. Considering the individual aptamers of the tandem riboswitches together with singletons of the two types, the authors observed that tandem aptamer 1 clustered with singleton type 1, and tandem aptamer 2 with singleton type 2. On a strictly structural level, the predicted structures of these two clusters also showed high similarity within the cluster. The singleton aptamers can also be grouped based on structure as more similar to the first or the second aptamer of the tandem. To add to the evidence supporting ghosts being the degraded versions of individual aptamers in the tandem, when looking at the secondary structure, the ghost aptamers appear similar to specific stem-loop regions of the corresponding aptamer in the tandem as well.
To evaluate the universality of the approach, the authors also repeated their network-based clustering analysis of singleton and tandem aptamers, and observe similar patterns of sequence conservation, first in Actinobacteria, and then in a much more diverse randomly sampled sequence set spanning multiple phyla.
These results suggest that the tandem aptamer came first evolutionarily, and the singleton aptamers present in different bacteria are descendants of the tandem ancestor. Which aptamer will be more conserved appears to be dependent on what gene is being regulated by the riboswitch (Figure 1).
Why I Find this Work Exciting
This study is a diligent and systematic look at riboswitches across many bacterial families, and resolves a seemingly contradictory set of results obtained in two different species.
The network-based sequence similarity comparisons are not limited by the type of metric used. In fact, the authors use a number of different sequence similarity metrics.
For biological sequences it might be more intuitive to think about network-based clustering that would connect divergent sequences with lots of intermediates connecting them. Classical phylogenetic approaches apply hierarchical clustering, which also groups sequences based on similarity, but might not account for such “reachability”, especially when dealing with short sequences, with lots of primary sequence variability, but constrained by secondary structure.
My absolute favorite part of this study are the ghosts – the remnants of individual aptamers that accompany singletons. They were described previously as being relevant to stabilizing the structure of the singlet riboswitches, but it was not clear whether the singlet glycine riboswitches preceded the tandem arrangement in their evolution, or vice-versa. This study by Crum et al. points to the nomenclature “ghost” aptamer also being correct, as it suggests the tandem glycine riboswitch was the ancestral form, and the ghost aptamers are what remain of the less conserved aptamer in different contexts.
Future Directions and Questions for the Authors
In the discussion, the authors mention that this graph clustering approach can be used with other riboswitches or ncRNA. Would it also be a reasonable approach to study the evolution of fast evolving viral genomes or any other organism/genetic element?
Are there regions/nucleotides that might be covarying with the regulated gene itself? Or covarying with some other part of the genome?
Are there ways of predicting whether a riboswitch will be an on-switch or an off-switch in silico?
The authors alluded to horizontal gene transfer as a potential explanation as to why the TP-regulating Bacillaceae riboswitches are more similar to TP-regulating Vibrionaceae ones. If this is the case, has the downstream gene been transferred together with the regulatory elements? Or is it just the riboswitch that might have been exchanged between taxa?
References
Babina, A.M., Lea, N.E., Meyer, M.M., 2017. In Vivo Behavior of the Tandem Glycine Riboswitch in Bacillus subtilis. mBio 8, e01602-17. https://doi.org/10.1128/mBio.01602-17
Bayegan, A.H., Clote, P., 2018. RNAmountAlign: efficient software for local, global, semiglobal pairwise and multiple RNA sequence/structure alignment. bioRxiv 389312. https://doi.org/10.1101/389312
Ruff, K.M., Strobel, S.A., 2014. Ligand binding by the tandem glycine riboswitch depends on aptamer dimerization but not double ligand occupancy. RNA 20, 1775–1788. https://doi.org/10.1261/rna.047266.114
Posted on: 30 October 2019
doi: https://doi.org/10.1242/prelights.14930
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioinformatics category:
Expressive modeling and fast simulation for dynamic compartments
Transcriptional profiling of human brain cortex identifies novel lncRNA-mediated networks dysregulated in amyotrophic lateral sclerosis
Spatial transcriptomics elucidates medulla niche supporting germinal center response in myasthenia gravis thymoma
Also in the evolutionary biology category:
A long non-coding RNA at the cortex locus controls adaptive colouration in butterflies
AND
The ivory lncRNA regulates seasonal color patterns in buckeye butterflies
AND
A micro-RNA drives a 100-million-year adaptive evolution of melanic patterns in butterflies and moths
A complex Plasmodium falciparum cryptotype circulating at low frequency across the African continent
Evolution of tandem repeats in putative CSP to enhance its function: A recent and exclusive event in Plasmodium vivax in India
preLists in the bioinformatics category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Also in the evolutionary biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |











 (No Ratings Yet)
(No Ratings Yet)