Retrieving High-Resolution Information from Disordered 2D Crystals by Single Particle Cryo-EM
Preprint posted on 6 December 2018 https://www.biorxiv.org/content/early/2018/12/06/488403
Article now published in Nature Communications at http://dx.doi.org/10.1038/s41467-019-09661-5
2D crystals revisited – High resolution structures of the MloK1 ion channel are solved from “imperfect” 2D crystals by treating them as a single particle cryoEM sample
Selected by David WrightCategories: biochemistry, biophysics, molecular biology
Background
Membrane proteins, despite making up over half of small molecule drug targets (1), make up less than 2% of the Protein Data Bank (PDB). Historically, X-ray crystallography was the most popular technique for the solution of membrane protein structures, however cryogenic electron microscopy (CryoEM) is now the go-to technique for large, flexible membrane proteins. There are many reasons why cryoEM is particularly suited for membrane proteins, but one of the most important is that cryoEM copes particularly well with the multitude of conformations commonly found in a membrane protein sample. MloK1 is a homotetrameric prokaryotic ion channel, which is used in this preprint as a test case for membrane proteins more generally.
The very first membrane protein structures were generated from 2D crystals (2), formed from the proteins aligning side-by-side as if in a native bilayer. Due to the fact that there is only one protein molecule in the z-axis, the resolution in this plane is often very poor. With the resolution revolution (3) due to improved hardware and software, single particle cryoEM has emerged as a high resolution technique to rival X-ray crystallography. In single particle cryoEM of membrane proteins the sample is typically solubilised in detergent, which might lead to a non-native structure. On the other hand, 2D crystals are grown in a lipidic environment, so might result in a more native-like structure. This preprint uses novel software for single particle reconstruction of 2D crystals deemed “too imperfect” for traditional 2D electron crystallography.
Results
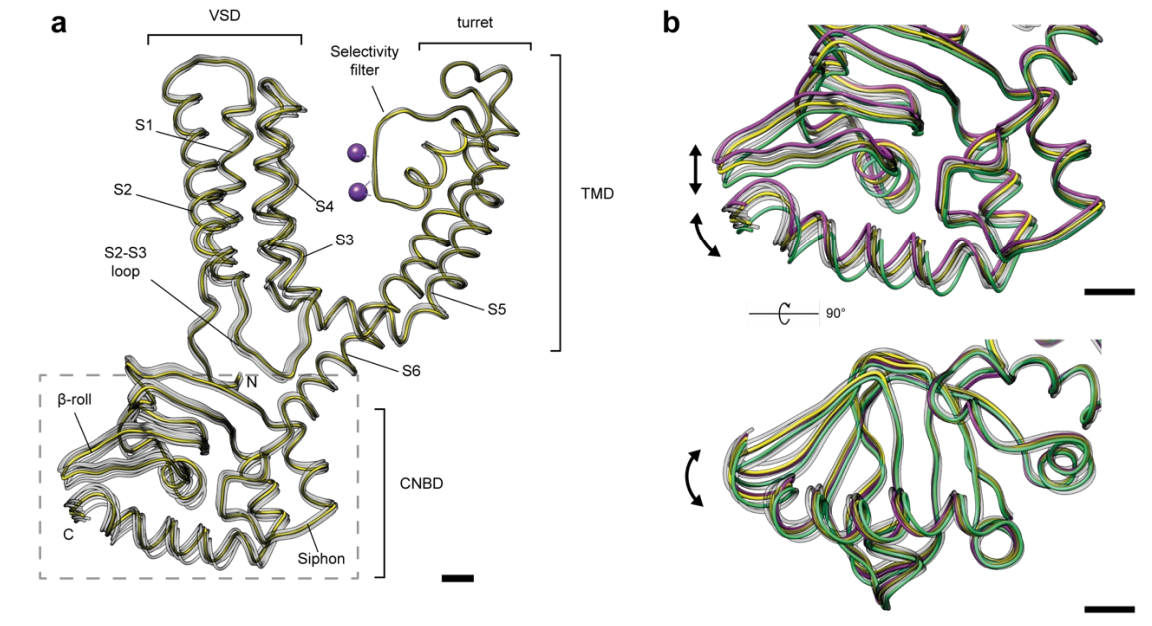
The first point worthy of note is that the overall resolution of the final model was 4 Å, which is higher than the previously available MloK1 structure (4). Further to this, the authors calculate the Brag reflections of these data to be 10 Å or worse, suggesting that this hybrid approach actually improves the resolution of the model generated from these data. It might be expected that the “single particle” selected in analysis was one tetramer; however it seems that there are at least 9 tetramers in each single particle. Even though this ion channel is homotetrameric, it might be possible for the structures to be asymmetric; however, using localized reconstruction, “no significant deviations from 4-fold symmetry” were detected.
The most interesting result, in my opinion, is that eight different structures are generated from one crystal system. If this were crystallography, we would expect a single “averaged” structure, much like the 4 Å structure described above. By generating 3D classes, however, the different conformations can be identified, resulting in structures to 4.4 – 5.6 Å resolution. The largest RMSD between states was less than 1 Å, suggesting that there were no vast global differences between states (this is illustrated in figure 4 included below of the preprint and can be visualised in supplementary movies 1-3). It was also noted that there was no correlation between structure and position on the crystal e.g. that state A was not always found on the edge of the crystal (see supplementary figure 8).
Conclusions and comments
This work resulted in a structure with a higher resolution than that previously seen with MloK1. 2D crystallography generally leads to a structure with good resolution in the x and y planes (as there are thousands of molecules here) but very poor resolution in the z plane (as only one molecule is present). This results in what is often known as the “missing cone” of data. As these crystals are not perfectly planar, the “single particles” are tilted relative to each other, which offers slightly different views i.e. not all from directly above. There is still significant missing data, but this technique, for imperfect crystals, is clearly an improvement on typical 2D crystallography. To the authors’ knowledge, this is the first example of 3D classification within a 2D dataset, and I think it will certainly not be the last. This technique could be very interesting for use in 2D arrays of membrane proteins that are sometimes natively observed, for example in mitochondria and chloroplasts.
Why I chose this preprint
This is a really exciting article where the authors rescue poorly ordered crystals and generate a really nice set of structures. It is really impressive that the imperfections in the crystal that led to non-planarity have been exploited to minimise the “missing cone” that usually affects resolution in the z-axis. Hopefully this work will be applicable to other projects that have stalled with poorly-diffracting 2D crystals. This workflow has also been incorporated into the FOCUS package for other users’ future benefit.
Outstanding questions
- How easy it is to make 2D crystals in reality? How much sample is required compared to standard single particle cryoEM?
- If purified protein is available, is there any benefit to forming 2D crystals over single particle cryoEM?
- How many examples of native 2D crystals are there and would this approach be able to handle crystals with multiple, potentially heterogeneous components?
References
- Santos R, Ursu O, Gaulton A, Bento AP, Donadi RS, Bologa CG, et al. A comprehensive map of molecular drug targets. Nature Reviews Drug Discovery. 2016 12/02/online;16:19.
- Henderson R, Unwin PN. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975 Sep 4;257(5521):28-32. PubMed PMID: 1161000. Epub 1975/09/04. eng.
- Kühlbrandt W. The Resolution Revolution. Science. 2014;343(6178):1443.
- Kowal J, Biyani N, Chami M, Scherer S, Rzepiela AJ, Baumgartner P, et al. High-Resolution Cryoelectron Microscopy Structure of the Cyclic Nucleotide-Modulated Potassium Channel MloK1 in a Lipid Bilayer. Structure. 2018 2018/01/02/;26(1):20-7.e3.
Posted on: 17 December 2018
doi: https://doi.org/10.1242/prelights.6331
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biochemistry category:
Structural basis of respiratory complexes adaptation to cold temperatures
Lens Placode Modulates Extracellular Matrix Formation During Early Eye Development
Generalized Biomolecular Modeling and Design with RoseTTAFold All-Atom
Also in the biophysics category:
Structural basis of respiratory complexes adaptation to cold temperatures
Actin polymerization drives lumen formation in a human epiblast model
Learning a conserved mechanism for early neuroectoderm morphogenesis
Also in the molecular biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Nanos2+ cells give rise to germline and somatic lineages in the sea anemone Nematostella vectensis
Plant plasmodesmata bridges form through ER-driven incomplete cytokinesis
AND
Plasmodesmata act as unconventional membrane contact sites regulating inter-cellular molecular exchange in plants
preLists in the biochemistry category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
Preprint Peer Review – Biochemistry Course at UFRJ, Brazil
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biochemistry deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the biophysics category:
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
66th Biophysical Society Annual Meeting, 2022
Preprints presented at the 66th BPS Annual Meeting, Feb 19 - 23, 2022 (The below list is not exhaustive and the preprints are listed in no particular order.)
| List by | Soni Mohapatra |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Biophysical Society Meeting 2020
Some preprints presented at the Biophysical Society Meeting 2020 in San Diego, USA.
| List by | Tessa Sinnige |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Biomolecular NMR
Preprints related to the application and development of biomolecular NMR spectroscopy
| List by | Reid Alderson |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
Also in the molecular biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (No Ratings Yet)
(No Ratings Yet)