Systematic functional analysis of Leishmania protein kinases identifies regulators of differentiation or survival
Preprint posted on 2 December 2020 https://www.biorxiv.org/content/10.1101/2020.09.06.279091v2
Article now published in Nature Communications at http://dx.doi.org/10.1038/s41467-021-21360-8
Categories: cell biology, molecular biology
Background
Many vector-borne pathogens have complex life cycles due to their requirement to transition between insect and mammalian hosts. In these pathogens, cell type differentiation is central to their ability to adapt to different environments. Some parasitic protozoa undergo cell cycle arrest in response to autocrine signals in their host, and can undergo differentiation in response to environmental cues to produce a different cell type that can proliferate. Although in general, little is known about the molecular mechanisms behind these events, the parasites Plasmodium, Trypanosoma brucei and Leishmania have begun to provide some insight into parasite differentiation and survival. Phosphorylation-mediated signal transduction likely plays a pivotal role in Leishmania differentiation, as previous studies have reported protein phosphorylation changes throughout the parasites’ various life cycle stages. Although various studies have explored the role of individual Leishmania protein kinases less than 10% of the kinome has been investigated by genetic and chemical approaches. In the present preprint, Baker et al (1) systemically tagged protein kinases with mNeonGreen fluorescent protein for localization studies, and generated null mutants using CRISPR-Cas9 to study Leishmania survival, differentiation and infection success in vitro and in vivo in both the invertebrate and vertebrate hosts.
Key findings and developments
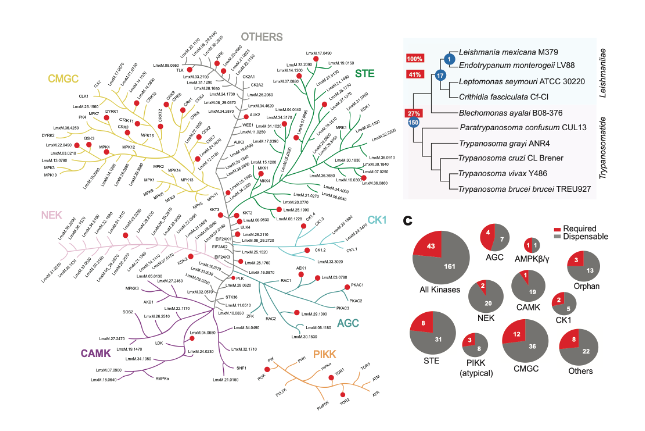
Generation of gene deletion mutants. The authors began by investigating 204 Leishmania mexicana protein kinases (193 eukaryotic protein kinases and 11 atypical protein kinases). From these, 174 were found to have orthologues in trypanosomes and Leishmania, while 17 were unique to Leishmaniinae (termed LUKs for Leishmaniinae unique kinases). Following identification, the authors attempted to generate gene deletion mutants using CRISPR-Cas9. Gene deletion mutants were successfully produced for 161 protein kinases (that is, 79% of the kinome), while 43 (21%) were found to be essential for promastigotes. 41% of LUKs were essential, which the authors argue is a suggestion that Leishmania promastigotes require these LUKs specifically for life cycle adaptations.
Localization of protein kinases. The authors generated 199 N- or C-terminal mNeonGreen-tagged protein kinases for localization studies in procyclic promastigotes. They used the atlas of Leishmania cellular landmarks and localized proteins to various cell compartments including the cytoplasm, basal body, nucleus, endomembrane, flagellum, lysosome, flagellar pocket, pellicular membrane, cytoplasmic organelles and mitochondrion. Interestingly, the fluorescence signal for some protein kinases varied during the cell cycle.
Phenotypic characterization of gene deletion mutants. The successful mutants were pooled and subjected to Leishmania life cycle progression. Mutants were tested using bar-seq analysis , for their ability to transition through the Leishmania life cycle including promastigotes, metacyclic promastigotes, axenic amastigotes, amastigotes in macrophages and amastigotes in the footpads of mice. The relative growth rate of each mutant was determined by counting the barcodes represented in each time point, and calculating the proportion of each mutant within the population. Outputs for each time point were defined as no loss of fitness, increased relative fitness or decreased relative fitness. Barcodes for each protein kinase were analysed individually, and as clusters, sorting mutants into groups with similar phenotypes taking all time points into account. The authors then used the projection pursuit method to calculate differences between each time point within the series for each of the mutant strains. Various clusters for each of the experimental arms were produced, revealing functional phenotypic groups of protein kinases involved in differentiation from metacyclic promastigote to amastigote, and growth and survival in macrophages and mice. To analyse colonization of the sand fly vector heatmaps were used to show relative loss of fitness, and motility mutants were analysed using transwell migration assays. The latter of which concludes that some of the proteins are fundamental to infection for a reason independent of flagellum defects.
The authors conclude that this unbiased interrogation of protein kinase function in Leishmania allows targeted investigation of organelle-associated signalling pathways required for successful intracellular parasitism.
What I like about this preprint
I have a great interest in parasites, and find the question being targeted in this study, a vital one for our understanding of parasitism. I think the findings in this study, which takes advantage of state-of-the-art molecular tools for phenotypic characterization, will be an interesting baseline for a lot of questions both specific to Leishmania, and general to parasitology.
References
- Baker N and Catta-Preta C, et al, Systematic functional analysis of Leishmania protein kinases identifies regulators of differentiation or survival, bioRxiv, 2020.
Posted on: 18 December 2020
doi: https://doi.org/10.1242/prelights.26550
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cell biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Structural basis of respiratory complexes adaptation to cold temperatures
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
Also in the molecular biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Nanos2+ cells give rise to germline and somatic lineages in the sea anemone Nematostella vectensis
Plant plasmodesmata bridges form through ER-driven incomplete cytokinesis
AND
Plasmodesmata act as unconventional membrane contact sites regulating inter-cellular molecular exchange in plants
preLists in the cell biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |
Also in the molecular biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (No Ratings Yet)
(No Ratings Yet)