Tension on kinetochore substrates is insufficient to prevent Aurora-triggered detachment
Preprint posted on 13 September 2018 https://www.biorxiv.org/content/early/2018/09/13/415992
What tension? Pulling forces in a proper bi-polar spindle are thought to turn off Aurora B-directed corrections. An in vitro study suggests that tension on kinetochores alone cannot deter this kinase
Selected by Angika BasantCategories: cell biology
Background:
When proliferating, cells divide their genetic material by employing a highly complex and dynamic machine – the mitotic spindle. Microtubules emanating from opposite poles of the spindle contact duplicated chromosomes at specialised macromolecular structures called kinetochores. Microtubule-kinetochore interactions are a vital aspect of cell division and have incidentally been the focus of many preprints quite recently (1-3).
To faithfully pull apart sister chromatids, i.e. to provide every new cell a full complement of genetic material, each kinetochore in a pair must attach to a microtubule from opposite poles. Crucially, cells must be able determine whether this has been achieved before segregating chromosomes. This complex task is performed by Aurora B kinase. Proper attachments involve kinetochore pairs pulled on from opposite poles, thereby generating tension on kinetochores. Aurora B selectively phosphorylates its substrate proteins on kinetochores under low tension (4), where presumably both kinetochores attach to one pole. This phosphorylation reduces their affinity for microtubules, destabilising these erroneous attachments. How does Aurora B sense the level of tension on kinetochores, and is that the only cue underlying kinase-triggered detachment of microtubules?
Kinetochores are known to undergo dramatic structural changes when under tension (5-7). An appealing hypothesis is that this occludes key Aurora B substrates from being phosphorylated, thus retaining correct microtubule attachments. It is a considerable challenge to test such models in vivo. Aurora B performs several roles in cell division and isolating its function at kinetochores, particularly in the presence of opposing phosphatase activity, is not trivial.
Key findings:
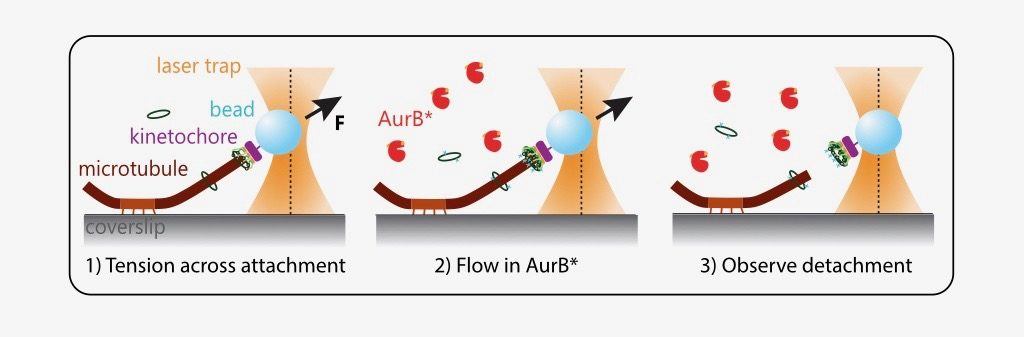
The authors in this study distil this complex intracellular phenomenon into a reconstituted in vitro assay. They purify yeast kinetochores such that endogenous kinase and phosphatase activity has been quenched. Next, they design and purify an active recombinant Aurora B kinase (AurB*) that can phosphorylate key microtubule-interacting substrates (particularly the Ndc80 protein) on isolated kinetochores.They show that AurB* phosphorylation impairs the ability of these kinetochores to bind fluorescently labelled, stabilised microtubules in vitro. This effect is partially Ndc80 dependent.
What is the effect of AurB* on physiologically relevant microtubule attachments that are load-bearing, end-on and dynamic? The authors modified an optical trap assay (8) to generate tension on microtubule-attached kinetochores where AurB* can be flowed in. In such experiments, kinetochores linked to polystyrene beads are added to coverslips on which anchored microtubules are grown. The position and force experienced by these beads can be controlled via a tightly focused laser beam (optical trap). The kinetochores are then indirectly manipulated by the laser to attach to tips of dynamic microtubules and held at a desired force (to generate tension). Exogenous AurB* is added once the attachment is made.
While these manipulations are technically challenging, the authors were able to observe that phosphorylation by AurB* releases tips of microtubules from kinetochores under low tension (~1 piconewton (pN)), as would be predicted for an improper attachment in vivo. A kinase-dead mutant of AurB* is 3-fold less effective in performing this “correction”.
Importantly, the prevailing model also predicts that a properly attached kinetochore under high tension should not be detached from microtubules by Aurora B. However, quite surprisingly, the authors find that AurB* activity releases microtubule attachments even at high tension regimes (5 and 8 pN). This suggests that tension across kinetochores is not sufficient to block Aurora B phosphorylation on kinetochore substrates.
What I like about this preprint:
It is very exciting to see a reconstituted system that draws on decades of work and has the potential to test several dogmas in the field of chromosome segregation. To quote the authors, “Rigorously testing such models requires independent control of tension, attachment, and enzyme activity”. This study (a) tackles all three of these challenges, (b) reveals unexpected complexities in the system and (c) sets the stage to further probe elusive mechanisms of how our genomes are faithfully divided.
Future directions and questions for the authors:
In the light of their data, the authors state that alternate models for error correction bear consideration. For example, we think of tension as primarily modifying the kinetochore but it may instead affect kinase activity or even that of a phosphatase. These would be very exciting new directions for the field. However, to delve further in the current experiments, I would love to ask the authors a few questions:
- How do detachment rates of non-phosphorylatable kinetochores (Ndc80-7A) compare with wild-type under low and high tension? In other words, could detachment under high tension be occurring via an unexpected mechanism?
- Could the response to kinetochore tension depend on strength of AurB activity? That is, if AurB* activity is too high, perhaps it phosphorylates its kinetochore substrates regardless of tension. In vivo AurB activity at kinetochores might be tightly regulated to respond to changes in tension. Were a range of AurB* concentrations tested, other than 0.5uM and 5uM, as even 0.5uM of exogenous, chimeric AurB* may exceed a threshold?
- It seems surprising that not only does AurB* function persist at high tension, but the detachment rates increase with increasing force. How would you explain this positive correlation?
References:
- Kuhn and Dumont, bioRxiv 2018; https://doi.org/10.1101/463471
- Roy et al., bioRxiv 2018; https://doi.org/10.1101/459594
- Doodhi et al., bioRxiv 2018; https://doi.org/10.1101/455873
- Kelly and Funabiki., Curr. Op. in Cell Biol 2009
- Wan et al., Cell 2009
- Joglekar et al., Curr. Biol 2009
- Tien et al., Genetics 2014
- Akiyoshi et al., Nature 2010
Posted on: 11 November 2018 , updated on: 12 November 2018
doi: https://doi.org/10.1242/prelights.5522
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cell biology category:
Fetal brain response to maternal inflammation requires microglia
Alteration of long and short-term hematopoietic stem cell ratio causes myeloid-biased hematopoiesis
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
preLists in the cell biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |











 (No Ratings Yet)
(No Ratings Yet)