The context-dependent, combinatorial logic of BMP signaling
Preprint posted on 8 December 2020 https://www.biorxiv.org/content/10.1101/2020.12.08.416503v1
Article now published in Cell Systems at http://dx.doi.org/10.1016/j.cels.2022.03.002
How do ligand-receptor dependent signalling pathways acquire contextuality and specificity?
Selected by Meng ZhuCategories: biochemistry, molecular biology
Background:
Cells within a tissue communicate through signalling molecules. An important first step of this communication is the interaction between secreted signaling molecules, also called ligands, and receptors on the cell surface. Cell-cell signaling can be classified into groups of ligands and receptors that together activate the same downstream effects. Well-studied examples of these include the Fibroblast Growth Factor (FGF), Wnt, and Bone Morphogenetic Protein (BMP) pathways. These pathways are functionally important in developmental, neural, and physiological contexts. In development, for example, interaction between ligands and receptors of these pathways is key to cell proliferation, growth, and differentiation, and the correct expression of these ligands and receptors in certain spatial and temporal patterns is key to embryonic patterning.
A fundamental feature of these signalling pathways is redundancy of signaling components – the genome of a multicellular animal can encode several ligands and receptors belonging to the same signalling pathway, and these members are often co-expressed in a certain combination for the various developmental stages or tissues where the pathway plays a role. As inferred by previous studies, a pathway’s many ligand variants can promiscuously bind nearly any receptor variant to activate the pathway, yet, in certain cell contexts, the ligand variants appear to have non-redundant roles. These properties contribute to the complexity of signalling interactions, but also raise several questions relating to the logic of signalling regulation given a certain ligand-receptor expression pattern. For example, can sequence homology between ligands or receptors determine their functional redundancy? And to what extent can different ligands and receptors replace each other? Addressing these questions can not only advance our understanding of signalling pathway interactions, but also help us to better interpret the expression profile of members of a certain signalling pathway in a specific tissue.
In this preprint, Klumpe et al. focused on the BMP pathway as an example to address these questions of signalling specificity. They used a systematic and quantitative approach to survey how combinations of ligands activate the BMP pathway and how these combinatorial properties vary between cell types. Using these data, they can compare the similarity of each ligand’s signaling activity and ask if cells perceive the same differences between ligands, or not. Finally, they used a mathematical model and simulated receptor-ligand binding to provide putative explanations for the functional similarities between different ligands and receptors.
Key findings:
To quantitively probe the combinatorial interactions between BMP ligands, the authors constructed a BMP reporter NMuMG cell line by hooking a Smad1/5/8 responsive element (which activates downstream transcription when the upstream BMP pathway is activated), with a fusion Histone 2B (H2B)-mCitrine. The authors used this reporter line to first determine the individual behavior of 10 BMP ligands when activating BMP receptors expressed in NMuMG cells (ACVR1, ACVR2A, ACVR2B, BMPR1A, and BMPR2), and then moved on to quantify pairwise responses. The parameters that they initially focused on were half-maximal activation (EC50) and Relative Ligand Strength (RLS), which measures pathway activity at saturating concentrations normalized to the activity of the strongest ligand. However, the authors found that the ligands that display similar RLS can display differences in the way they interact with other ligands. To better evaluate the ligand equivalence, the authors defined a new parameter, the so- called interaction coefficient (IC), which compares the differences between the responses to a ligand when combined with itself or with another ligand. Based on the IC regime, the interactions between ligands in a pair can be classified into “saturated additive” (IC= 0), “negative interactions” (IC<0), and “positive synergy” (IC>1) (Figure).
The authors found that most pairs fell into the “saturated additive” category, whereas others ranged from antagonism to suppression, two kinds of negative interactions (IC < 0). Based on the interaction pattern, the BMP ligands can be grouped into [BMP2, BMP4], [BMP5, BMP6, BMP7, BMP9], [BMP10], [GDF5, GDF7], and [GDF6] (Figure).
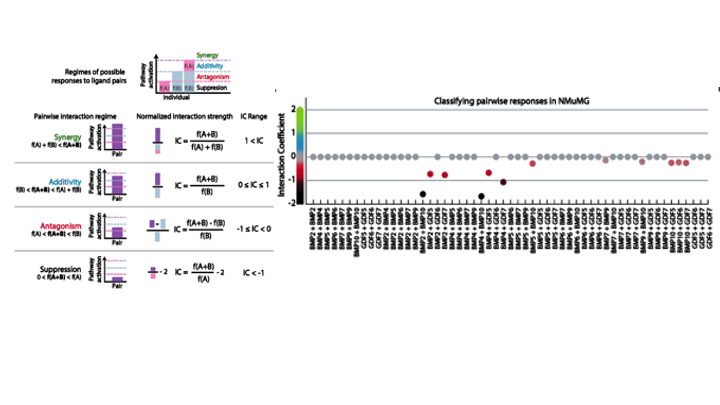
Figure. The definition of Interaction Coefficient (IC) and pairwise analysis of BMP ligands based on IC.
To determine to what extent the interaction metric identified in the NMuMG cells holds true for other cell types, the authors applied the same approach to assay BMP ligand interactions in another cell type, the mouse embryonic stem cell (mESC). The authors found significant differences in single ligand binding parameters as well as pairwise interactions between different ligands. These results thus suggest that ligand interactions are context-dependent.
What leads to the contextuality? The authors hypothesized receptor expression to be a cause, as NMuMG and mESC cells differ significantly in their BMP receptor expression profiles. To address this, they first downregulated ACVR1 in NMuMG cells to make its expression level more similar to that of the mESCs. They found that this allows the narrowing of certain differences in the interaction metric between NMuMG and mESC cells, but not others. The authors then downregulated the two most highly expressed receptors, BMPR1A and BMPR2. Knocking down BMPR2, similar to ACVR1 knockdown, recapitulates a difference between NMuMG and mESC and generates some aspects of ligand equivalence observed in mESC. These results suggest that the expression of receptors can account to a certain extent for the how ligand interactions change between different cell types. Besides downregulation of highly expressed receptors, the authors also performed experiments to ectopically express the two lowest expressed receptors, ACVRL1 and BMPR1B. The authors found that the upregulation of these two receptors allows the removal of many of the suppressive effects of their primary binding ligands. These results suggest that the non-additive interactions, such as antagonism and suppression, may arise from receptor competition.
To better understand the ligand equivalence, the author generated a more global ligand equivalence picture by combining datasets for different cell types and clustered the BMP receptors as 5 equivalent categories [BMP2, BMP4], [BMP5, BMP6, BMP7], [BMP9], [BMP10], and [GDF5, GDF6, GDF7]. By analysing the protein sequence, the authors found that, surprisingly, the ligand sequence similarity does not fully predict functional similarity in a single receptor context, though it did match the global functional similarity across all cell lines. Then what can account for the functional similarity, or ligand equivalence in general, and how it changes across contexts? The authors turned to a mathematical approach to simulate receptor-ligand binding scenarios. The model takes into account primarily the affinities to form a trimeric ligand-receptor complex and the limitation of receptor amount to generate ligand competition, and the level of activity for each type of complex. The parameters of the model were fitted using experimental measurements coming from individual and pairwise analysis performed above.
With best fit parameters, the model can recapitulate many of the experimental observations on differential behaviours of ligand groups, suggesting that these experimentally observed behaviours can be explained by a model of affinity-based competition. Furthermore, and more importantly, the model predicted the generation of high activity and low activity complexes between different combinations of BMP receptors and ligands. The low ligand activity is associated not with the formation of fewer complexes, but with the formation of many low activity complexes. The model also suggested that the change in BMP signalling by the change of single-receptor expression can result from direct or indirect effects as the receptor participates in the formation of a broad range of complexes. Finally, the model predicted that the addition of a second ligand can result in a shift in the activity of the first ligand, as the ligands influence each other’s ability to form complexes from shared components.
Overall, the study suggests several contextual mechanisms of which ligand equivalence to be controlled: 1) the expression of receptors, such as when BMP10 can only replace BMP9 in the presence of ACVRL1; 2) the composition of co-expressing ligands, such as when BMP4 and BMP7 can only replace each other in the absence of antagonising ligands such as GDFs. In general, this work addressed the mechanism of contextuality and is insightful for the studies of systems in which multi-component signalling pathways participate.
What I like about this preprint:
FGF, Wnt, and BMP pathways are the key players in a broad range of developmental contexts. They come on stage at different space and time to execute distinct functions. How these distinct functions can arise by the same category of signalling pathway is an important topic that I, as a developmental biologist, very much wish to understand. This work provides a systematic investigation to this matter and provides profound insights into this question. A take-home message of this work is that the identity of a ligand and receptor that is present in a certain context is crucial, and thus one cannot simply use the number of ligands or receptors in a certain signalling pathway category as a readout for the level of activity for this pathway.
Posted on: 11 February 2021
doi: https://doi.org/10.1242/prelights.27355
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biochemistry category:
Structural basis of respiratory complexes adaptation to cold temperatures
Lens Placode Modulates Extracellular Matrix Formation During Early Eye Development
Generalized Biomolecular Modeling and Design with RoseTTAFold All-Atom
Also in the molecular biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Nanos2+ cells give rise to germline and somatic lineages in the sea anemone Nematostella vectensis
Plant plasmodesmata bridges form through ER-driven incomplete cytokinesis
AND
Plasmodesmata act as unconventional membrane contact sites regulating inter-cellular molecular exchange in plants
preLists in the biochemistry category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
Preprint Peer Review – Biochemistry Course at UFRJ, Brazil
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biochemistry deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the molecular biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |











 (No Ratings Yet)
(No Ratings Yet)