The enteric nervous system of the human and mouse colon at a single-cell resolution
Preprint posted on 4 September 2019 https://www.biorxiv.org/content/10.1101/746743v4
Meet the 1%: single-cell RNAseq of human & mouse enteric nervous system reveals core transcriptional programs of enteric neurons, DEGs by circadian phase & colon location, new models for intestinal peristalsis, neuro-immune interactions, and more!
Selected by Jessica XieCategories: genomics
Background
Expanding upon the growing interest in traditionally understudied cell types, the latest publication from the Human Cell Atlas consists of an extensive single-cell RNAseq study of the enteric nervous system.
The enteric nervous system (ENS) is the most complex collection of neurons and glia in the body outside of the central nervous system (CNS). Innervating the gastrointestinal tract, the ENS controls and modulates many aspects of digestive function, such as intestinal motility, blood flow, nutrient absorption, as well as secretion of enzymes, ions, and many other substances. Moreover, it can do so entirely independently of the CNS, and therefore has been termed the “second brain”. While much less is understood about the interaction between the neurological and gastrointestinal system than the two systems in isolation, the ENS has received increasing attention of late for a number of reasons: a renewed emphasis on systems biology, the development of unbiased -omic techniques, and the ENS’s potential implication in several major diseases, such as inflammatory bowel disorder, Parkinson’s disease, and autism spectrum disorders.
Methods
The technical challenges that made this work a virtual impossibility until now are plain to see:
To begin, despite being a major part of the nervous system, ENS cells are a rare population (<<1%) in the colon. This necessitated the development of transgene mouse models to enrich for cell-types of interest prior to sequencing. Using fluorophores driven by the Sox10 and Uchl1 promoters to label early neural crest and mature neurons respectively [validated in Fig. 2], the authors eventually profiled 2,447 mouse enteric neurons and 2,710 glia. For the second part, the brute-force sequencing of an enormous number of cells had to suffice for human tissue: from >160,000 single nuclei, 831 human enteric neurons and 431 Interstitial Cells of Cajal (ICC) were identified.
The authors also performed extensive optimization of the nuclei extraction protocol [Fig. 3], after finding that snRNA-seq protocols previously applied to brain tissues adapted poorly to colon. Sequencing over 5,000 nuclei across 36 extraction conditions, they systematically varied several factors—
- detergent (NP40, CHAPS, Tween, or Digitonin),
- detergent concentrations,
- buffer (HEPES, Tris, Tricine),
- mechanical extraction conditions (dounced, chopped, or ground tissue),
- modifiers for use in nuclei isolation (salts, polyamines)
—and identified two conditions that recovered high-quality transcriptomic profiles from ENS cells with greatest efficiency and specificity. Electron micrography revealed that the first set of conditions (“TST”: 0.03% Tween-20 detergent, salts present, Tris buffer) isolated nuclei along with rough ER and attached ribosomes, while the second (“CST”: 0.49% CHAPS detergent, salts present, Tris buffer) isolated the nucleus, nuclear envelope, and ribosomes on the outer nuclear membrane. This second method, which was chosen as it captured more neurons with fewer contaminants, was subsequently given the catchier name “Ribosomes And Intact SIngle Nucleus (RAISIN)-seq”.
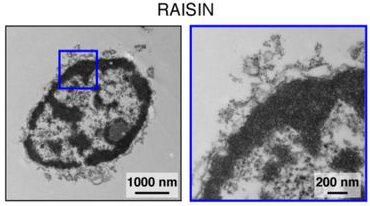
Results
a) Defining cell-types in the mouse & human ENS
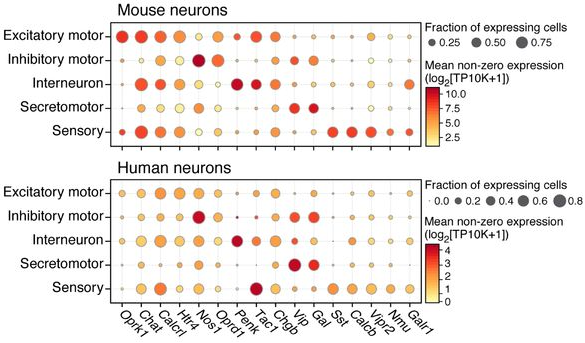
The 2,447 mouse and 831 human neurons were clustered into 24 and 11 subsets respectively, broadly consisting of similar groups [Fig. 6D, above]. Most abundant were cholinergic putative excitatory motor neurons (PEMNs) and nitrergic putative inhibitory motor neurons (PIMNs). Also identified were clusters of putative interneurons (PINs), putative sensory neurons (PSNs), and putative secretomotor/vasodilator neurons (PSVNs). Additionally, by matching mouse and human orthologs of differentially expressed genes that best distinguished each group, the authors identified a “core transcriptional signature” of each of these five major neuron groups, consisting of about 50–90 genes.
b) Factors affecting ENS cell composition and gene expression
The study design also enabled the authors to investigate how cell composition and gene expression in the murine ENS varied according to the following factors:
- age (from 11 to 52 weeks),
- sex (males and females),
- circadian rhythm phase (morning and evening),
- anatomical location (4 segments from proximal to distal colon).
First, batch correction was performed to remove the gene expression variation originally observed between different individual mice (or humans). The authors subsequently found that age and sex did not have much effect on gene expression.
On the other hand, significant gene expression changes were associated with circadian phase (e.g. upregulation of genes associated with cytoskeletal function, neuronal signaling, and neuro-immune signaling in the morning) and anatomical localization (e.g. higher expression of neurotransmitter receptors in distal mouse colon) [Fig. 5A & B]. Anatomical location affected cellular composition as well—for example, proximal colon was found to contain higher proportions of sensory neurons.
c) Functional insights
In addition to providing in-situ validation of their RNAseq results, the authors of this study also made valiant attempts to derive functional insights from their gene expression data—no easy task as the data, while substantial, was nonetheless inherently descriptive only.
For instance, the authors proposed an alternative model of the peristaltic reflex based on their motor neuron gene expression data. One previous model posited that enterochromaffin cells in the mucosa produce serotonin, which acts on sensory neurons, which in turn act on interneurons, which then activate motor neurons to stimulate peristalsis (muscle contraction and relaxation). However, this was inconsistent with some observations, for example that the mucosa was not necessary for contraction, and that serotonin release did not precede muscle contraction.
Here, the authors highlighted that almost all (cells putatively identified as) motor neurons expressed the Piezo-class mechanoreceptors, and may thus be able to directly sense distension. The authors further identified expression of a serotonin receptor in some sensory and motor neuron subsets, and on that basis suggested a more modulatory role for mucosa-derived serotonin.
The authors also put forth an alternative model of Interstitial Cells of Cajal (ICC) function; these are pacemaker cells that rhythmically regulate smooth muscle excitability. Previous models had proposed two types of roles for ICCs: a direct one, receiving signals from enteric neurons and transmitting these to smooth muscle; or a modulatory one, facilitating the direct signaling between neurons and smooth muscle.
The authors showed here that ICCs express the receptor for nitric oxide, the inhibitory neurotransmitter that directs muscle relaxation, while smooth muscle cells express the receptor for excitatory acetylcholine, which triggers muscle contraction. Based on this data, the authors proposed a third model that splits the difference: excitatory enteric neurons directly signal to smooth muscle to activate muscle contraction, while inhibitory enteric neurons signal to ICCs and induce muscle relaxation indirectly.
d) Some potential future directions
The authors concluded their study by inspecting their data from two other angles, presenting possible directions for future work.
First, they provided a preliminary description of cell-cell interactions of the ENS in Figure 7 by evaluating the expression of cognate ligand-receptor pairs in different cell-types, and through this identified several potential neuronal interactions with smooth muscle, with stromal adipocytes and fibroblasts, and also with immune cells.
Second, they showed in Figure 8 that several genes associated with ENS-related diseases—including autism, Parkinson’s disease, and intestinal bowel disease—were expressed in enteric neurons, opening the door to future investigations into their roles in disease.
Questions for the authors
Most unexpected finding? There were several scattered mentions throughout the paper, not all of which made it into this writeup—for example the findings that the serotonin synthesis enzyme Tph2 was not expressed, that some populations of sensory neurons may not arise from the Sox10 lineage, and that mice have greater sensory neuron diversity than humans—any you’d like to highlight in particular?
On that note, most exciting finding/ the one you’re most eager to follow up on?
Posted on: 7 October 2019
doi: https://doi.org/10.1242/prelights.14063
Read preprintHave your say
Sign up to customise the site to your preferences and to receive alerts
Register hereAlso in the genomics category:
Temporal constraints on enhancer usage shape the regulation of limb gene transcription
Transcriptional profiling of human brain cortex identifies novel lncRNA-mediated networks dysregulated in amyotrophic lateral sclerosis
A long non-coding RNA at the cortex locus controls adaptive colouration in butterflies
AND
The ivory lncRNA regulates seasonal color patterns in buckeye butterflies
AND
A micro-RNA drives a 100-million-year adaptive evolution of melanic patterns in butterflies and moths
preLists in the genomics category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Semmelweis Symposium 2022: 40th anniversary of international medical education at Semmelweis University
This preList contains preprints discussed during the 'Semmelweis Symposium 2022' (7-9 November), organised around the 40th anniversary of international medical education at Semmelweis University covering a wide range of topics.
| List by | Nándor Lipták |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)
4 years
Seema Khurana
Since you have the single cell RNA-Seq data, can you tell me if Advillin mRNA is expressed in these neurons.