The hourglass model of evolutionary conservation during embryogenesis extends to developmental enhancers with signatures of positive selection
Preprint posted on 2 November 2020 https://www.biorxiv.org/content/10.1101/2020.11.02.364505v1
Article now published in Genome Research at http://dx.doi.org/10.1101/gr.275212.121
Categories: bioinformatics, developmental biology
Introduction
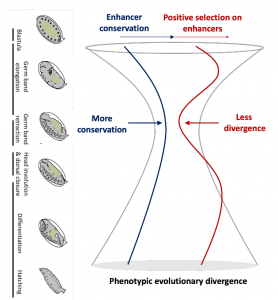
Since the 19th century there have been many attempts to find a pattern when comparing morphological trajectories of different species during development. In the 1990s the Hourglass (HG) model was proposed (Duboule 1994; Raff 1996). According to the HG model we should find, when comparing the morphology of animals in the same phylum, high variance in early and late development but low variance in mid-development. The conserved mid-developmental stage is usually referred to as the phylotipic stage or phylotypic period.
Although initially proposed based on morphology, the HG model has received support from many comparative gene expression studies. These studies found that the transcriptome of different species (e.g. Drosophila flies) is more similar in mid embryonic development (Kalinka 2010). Some recent analyses have found that the protein coding sequence of genes expressed in the phylotypic stage show relatively low positive selection and strong puryfing selection (Liu 2018 , Coronado Zamora 2019). However, it is still unknown if selection signatures on regulatory regions would follow this pattern.
Liu and collaborators tackle this question using DNAase-seq to identify stage specific enhancers throughout the embryonic development of the fruitfly, investigating their conservation and looking for signatures of positive selection.
About the preprint
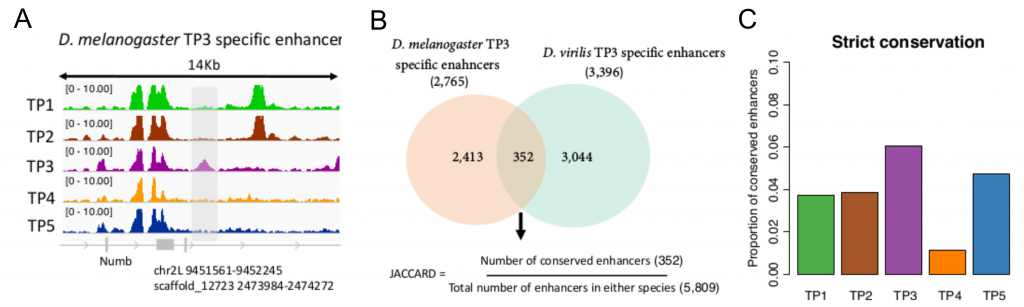
The authors performed DNase-seq to identify active regulatory elements across five matched embryonic development stages in two Drosophila species: D. melanogaster and D. virilis. Importantly, the five stages (TP1-TP5) include early, mid and late development, with stage TP3 within the phylotypic period.
Using a measure of enhancer conservation they found that TP3 (phylotypic period) has a higher proportion of conserved enhancers (Figure 1C). A high conservation in the sequence of enhancers can be, however, caused by strong purifying selection (in which most mutations are eliminated due to deleterious effects) or by a relatively weaker positive selection.
To test the latter scenario, they used a new approach that detects signatures of positive selection on genomic regulatory regions (Liu 2020). This approach is based on a Machine Learning algorithm that estimates the effect of nucleotide substitutions on chromatin stability (Gandhi 2014). More specifically, it consists of a gapped k-mer support vector machine (gkm-SVM) model. Support vector machine models are supervised learning models with associated learning algorithms that analyse data used for classification and regression analysis. Given a set of training examples, each marked as belonging to one or the other of two categories, an SVM training algorithm builds a model that assigns new examples to one category or the other. In this case the positive categories are stage-specific enhancers and the negative set is an equal number of sequences randomly sampled from the genome with similar characteristics (e.g. sequence length, nucleotide composition, etc). This gkm-SVM not only differentiates enhancer sequences from random sequences, but also identifies sequence features within the enhancer elements that determine their stage-specific occupancy.
Testing for positive selection
To identify signatures of positive selection, the authors used for further analyses only stage-specific enhancers with at least two substitutions between D. melanogaster and D. simulans. D. simulans is evolutionary closer to D. melanogaster (2.5 MY divergence) than D. virilis (40 MY divergence), being therefore a better option for this approach.
Using D. yakuba as an outgroup, they inferred the enhancer ancestor sequence and then calculated a deltaSVM score, which estimates the effect of substitutions on the gkm-SVM function score. This means that, if the substitutions that occurred in the D. melanogaster branch after divergence from D.simulans increased the chromatin accessibility of an enhancer, the deltaSVM score of this enhancer would be positive, and if the score is greater than expected by chance, it is assumed to have been positively selected.
The authors found that the phylotypic period (stage TP3) had a much lower proportion of enhancers with evidence of positive selection than the other stages. The pattern of conservation and positive selection on developmental enhancers seems then to support the Hourglass model (Figure 2).
Why I chose this preprint
Until now, the Hourglass model has been mostly tested using developmental transcriptomics data. This is the first study that tests the Hourglass model at the regulatory level trying to elucidate the role of developmental enhancers in the evolution of gene expression during embryogenesis. With this study, the authors help filling in a knowledge gap in the HG field.
Questions to the authors:
Q1: Instead of “U” shape expected from the HG model, it seems that the positive selection on enhancers shows an “M” shape. What do you think would be the cause for the relatively lower positive selection of the earliest and latest stages you analysed?
Q2: Do you think selective pressures on developmental processes could equally prevent positive selection on enhancers and codifying regions?
Q3: You mention that the higher number of enhancer in the phylotypic period suggests more redundancy and thus higher regulatory robustness in gene expression. Would it be possible to test this experimentally?
References
Duboule D. 1994. Temporal colinearity and the phylotypic progression: a basis for the stability of a vertebrate Bauplan and the evolution of morphologies through heterochrony. Development 1994:135–142.
Raff RA. 1996. The shape of life : genes, development, and the evolution of animal form. University of Chicago Press.
Kalinka AT, Varga KM, Gerrard DT, Preibisch S, Corcoran DL, Jarrells J, Ohler U, Bergman CM, Tomancak P. 2010. Gene expression divergence recapitulates the developmental hourglass model. Nature 468:811–814.
Liu J, Robinson-Rechavi M. 2018a. Developmental Constraints on Genome Evolution in Four Bilaterian Model Species. Genome Biol. Evol. 10:2266–2277.
Coronado-Zamora M, Salvador-Martínez I, Castellano D, Barbadilla A, Salazar-Ciudad I. Adaptation and Conservation throughout the Drosophila melanogaster Life-Cycle. Genome Biol Evol. 2019;11(5):1463-1482. doi:10.1093/gbe/evz086
Liu J, Robinson-Rechavi M. 2020. Robust inference of positive selection on regulatory sequences in human brain. bioRxiv. https://doi.org/10.1101/2020.03.09.984047.
Ghandi M, Lee D, Mohammad-Noori M, Beer MA. 2014. Enhanced Regulatory Sequence Prediction Using Gapped k-mer Features. PLoS Comput. Biol. 10:e1003711.
Posted on: 11 December 2020
doi: https://doi.org/10.1242/prelights.26321
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioinformatics category:
Expressive modeling and fast simulation for dynamic compartments
Transcriptional profiling of human brain cortex identifies novel lncRNA-mediated networks dysregulated in amyotrophic lateral sclerosis
Spatial transcriptomics elucidates medulla niche supporting germinal center response in myasthenia gravis thymoma
Also in the developmental biology category:
Temporal constraints on enhancer usage shape the regulation of limb gene transcription
OGT prevents DNA demethylation and suppresses the expression of transposable elements in heterochromatin by restraining TET activity genome-wide
Actin polymerization drives lumen formation in a human epiblast model
preLists in the bioinformatics category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Also in the developmental biology category:
GfE/ DSDB meeting 2024
This preList highlights the preprints discussed at the 2024 joint German and Dutch developmental biology societies meeting that took place in March 2024 in Osnabrück, Germany.
| List by | Joyce Yu |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
2nd Conference of the Visegrád Group Society for Developmental Biology
Preprints from the 2nd Conference of the Visegrád Group Society for Developmental Biology (2-5 September, 2021, Szeged, Hungary)
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
EMBL Conference: From functional genomics to systems biology
Preprints presented at the virtual EMBL conference "from functional genomics and systems biology", 16-19 November 2020
| List by | Jesus Victorino |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Society for Developmental Biology 79th Annual Meeting
Preprints at SDB 2020
| List by | Irepan Salvador-Martinez, Martin Estermann |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EDBC Alicante 2019
Preprints presented at the European Developmental Biology Congress (EDBC) in Alicante, October 23-26 2019.
| List by | Sergio Menchero et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Young Embryologist Network Conference 2019
Preprints presented at the Young Embryologist Network 2019 conference, 13 May, The Francis Crick Institute, London
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)