The modular mechanism of chromocenter formation in Drosophila
Preprint posted on 28 November 2018 https://www.biorxiv.org/content/early/2018/11/28/481820
Article now published in eLife at http://dx.doi.org/10.7554/elife.43938
Not just junk DNA: Satellite DNA and their cognate binding proteins form chromocenters to gather up chromosomes in one nucleus
Selected by Maiko KitaokaCategories: cell biology, molecular biology
Background
Eukaryotic chromosomes commonly have regions of satellite DNA abundant in tandem repeats at centromeric and pericentromeric chromatin. Centromeric heterochromatin is well known to establish kinetochore function and allow for faithful chromosome segregation. However, the role of pericentric satellite DNA is less understood, particularly due to the lack of protein-coding and conservation across species. Previous studies have shown roles for specific contexts only, but whether there is a more central role for eukaryotic satellite DNA, especially given its abundance in the genome, is not known.
With this new preprint, Jagannathan, Cummings, and Yamashita follow up on their recent eLife study discussing the formation and role of chromocenters, or the bundling of multiple chromosomes, to maintain nuclear organization. They address several new questions, including how all Drosophila melanogaster chromosomes in a genome can be bundled in chromocenters, and how multiple satellite DNAs and their corresponding DNA binding proteins can modulate this process together.
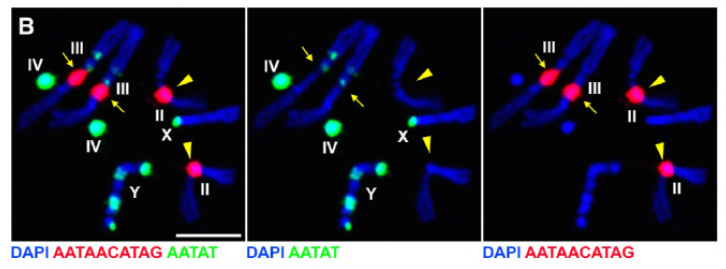
Key findings
Since their previous work identified a protein, D1, that was responsible for bundling chromosomes X, Y, and 4 into chromocenters, the group turned their attention to chromosomes 2 and 3, the major autosomes in D. melanogaster, to address how all chromosomes in the genome may be grouped into chromocenters. This led them to discover a longer satellite repeat on the autosomes that was bound by the Prod (proliferation disrupter) protein. Disrupting prod function led to micronuclei formation and increased DNA damage, ultimately resulting in cellular death. This was also seen in their previous work with D1 mutants. Interestingly, while D1 mutations affected the germline, prod mutations did not, thus suggesting that both act on different tissues in spite of their similar chromocenter-forming functions. However, D1 prod double mutants fail to develop past the embryonic stages and have increased micronuclei, establishing the essential requirement of chromocenter formation via satellite DNA.
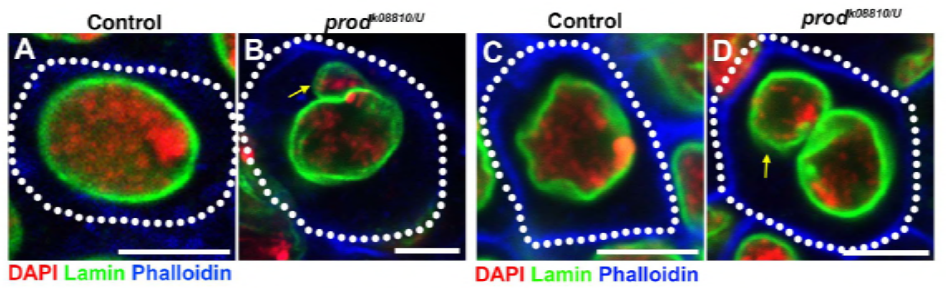
Prod clearly has a similar role to D1 in forming chromocenters. The authors expressed Prod ectopically in a tissue where it’s normally not present and caused the formation of chromatin threads that establish various chromosome territories connecting the autosomes. This clearly shows that Prod is sufficient to bundle these chromosomes together through its satellite DNA binding, explaining the mechanism of how chromocenters are formed.
At this point, Jagannathan and Cummings et al have established that Prod and D1 cluster chromosomes through satellite DNA. But since both proteins act on different sets of chromosomes, how is an entire genome’s set of chromosomes encapsulated in a nucleus? Prod and D1 did not seem to interact together through immunoprecipitation, suggesting only a weak or transient interaction. Through live imaging, the authors discovered a “kiss and run” interaction – D1 foci and Prod foci touch briefly and then separate, indicating a dynamic process of chromocenter formation. Both proteins seem to be mutually dependent on the other’s functional presence as well, since prod mutants showed defective D1 clustering in nuclei and vice versa. This interdependency provides a network to establish the bundling of all chromosomes in a genome.
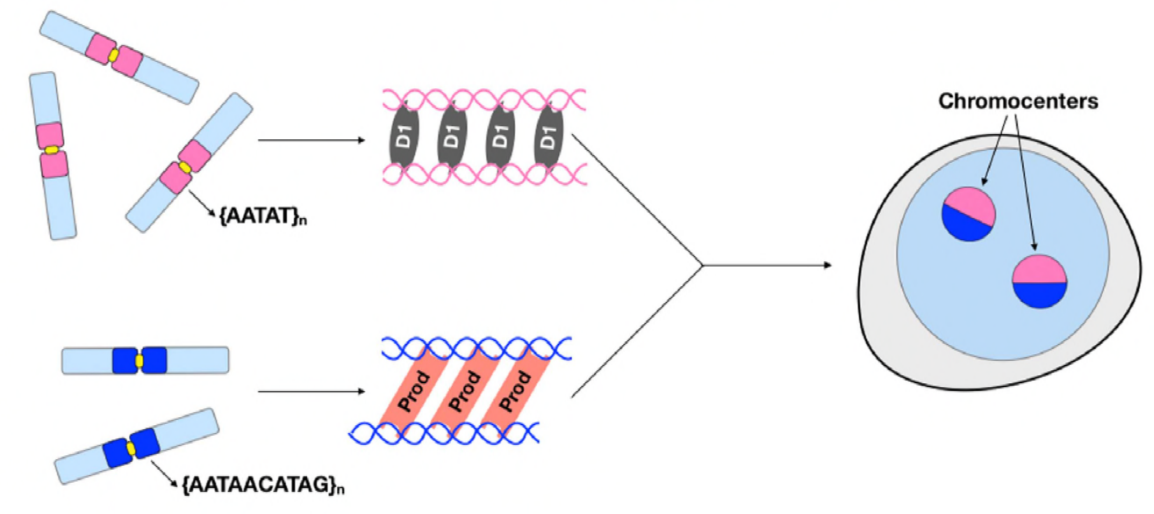
In summary, Jagannathan and Cummings et al have demonstrated that Prod and D1 create a network where both proteins bind their respective satellite DNA sequences in order to bring all chromosomes into a chromocenter, and eventually package the entire genome properly into the nucleus. Their study demonstrates the importance of satellite DNA, addressing not only the molecular and cell biological consequences of Prod and D1 perturbations but also the evolutionary significance of these proteins and satellite DNA as well.
Questions for the authors
The elife study showed that D1 loss causes micronuclei to bud off from the main nucleus – does prod loss cause micronuclei formation in the same way?
Do Prod and D1 interact with other proteins apart from their transient “kiss and run” interactions? Are these proteins known to function in other processes related to nuclear integrity and/or genome packaging?
In organisms with more than 4 chromosomes, how many chromocenter-forming, satellite DNA-binding proteins might be necessary to bundle a larger genome with more chromosomes?
The discussion mentions that D. simulans doesn’t have the satellite DNA that binds D. melanogaster Prod, so what is the Prod-like protein in D. simulans and how similar is it to D. melanogaster in structure and function?
Posted on: 11 December 2018
doi: https://doi.org/10.1242/prelights.6160
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cell biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Structural basis of respiratory complexes adaptation to cold temperatures
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
Also in the molecular biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Nanos2+ cells give rise to germline and somatic lineages in the sea anemone Nematostella vectensis
Plant plasmodesmata bridges form through ER-driven incomplete cytokinesis
AND
Plasmodesmata act as unconventional membrane contact sites regulating inter-cellular molecular exchange in plants
preLists in the cell biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |
Also in the molecular biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |











 (No Ratings Yet)
(No Ratings Yet)