The small RNA ErsA plays a role in the regulatory network of Pseudomonas aeruginosa pathogenicity in airways infection
Preprint posted on 23 June 2020 https://www.biorxiv.org/content/10.1101/2020.06.22.164558v2
Article now published in mSphere at http://dx.doi.org/10.1128/msphere.00909-20
ErsA in aeruginosa: a small RNA modulates the pathogenicity of P. aeruginosa in airway infections
Selected by Zhang-He GohCategories: immunology, microbiology, pharmacology and toxicology
preLight Author’s note
This is the third preLight of a three-part series on airway inflammation, infection, autophagy and its mediators. Links to the first and second preLights.
Background of preprint
ErsA is a small RNA (sRNA) in Pseudomonas aeruginosa that plays a key role in its response to stressful conditions during host infection. By interacting with other transcriptional factors, ErsA has been reported to modulate virulence traits like biofilm formation and motility [1-3], stimulate exopolysaccharide production [2], regulate the P. aeruginosa transcriptome [2], mediate the interaction between P. aeruginosa and its host [2], and even contribute to antibiotic resistance [4]. Given the prevalent role of ErsA in P. aeruginosa’s pathogenicity, Ferrara et al. assessed its role in respiratory infections caused by P. aeruginosa (Fig. 1). They showed that in acute infection, ErsA mediates regulation and contributes to the host inflammatory response; while in chronic infection, ErsA aids in adaptation and promotes the development of antibiotic resistance in P. aeruginosa.
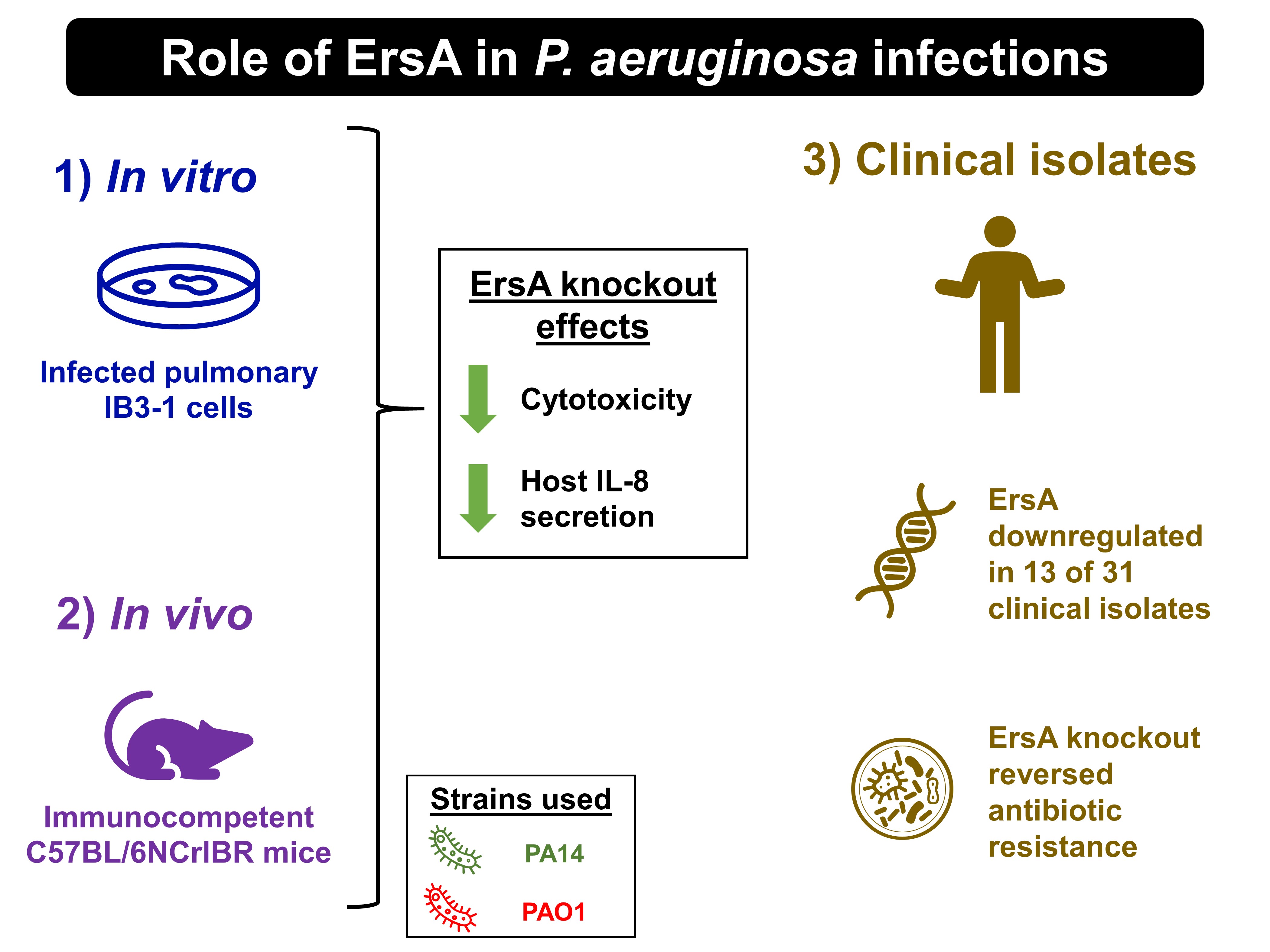
Figure 1. Role of ErsA in P. aeruginosa infections described by Ferrara et al..
Key findings of preprint
Ferrara et al. first evaluated ErsA’s role in regulating bacterial functions in cytotoxicity and stimulation of host inflammation in vitro. They infected pulmonary IB3-1 cells with two laboratory strains of P. aeruginosa, PA14 and PAO1, their knockout ErsA counterparts, and an ErsA-overexpressing PAO1 strain (pGM-ErsA). This experiment yielded two key observations. First, by comparing PA14 and PAO1 with their respective knockout ErsA counterparts, the authors showed that the loss of ErsA indeed attenuated the cytotoxic potential of P. aeruginosa. Second, IB3-1 cells infected with the PAO1 ErsA-knockout strain secreted less IL-8 than those infected with the PAO1 wild-type.
Having showed that ErsA enhances cytotoxicity and induces a pro-inflammatory response in host cells in vitro, the authors then infected immunocompetent C57BL/6NCrlBR mice with the PAO1 and PAO1 ErsA-knockout strains. Knocking out ErsA also decreased the mortality and attenuated the inflammatory response in mice acutely infected with the PAO1 strain. In contrast, neither the incidence of chronic colonisation in surviving mice nor the inflammatory response differed significantly between the PAO1 and PAO1 ErsA-knockout strains.
To determine whether the ErsA gene and its expression could be conserved independently of the origin of P. aeruginosa isolates, and whether patho-adaptive mutations are responsible for ErsA downregulation, the authors assessed the expression levels of ErsA in 31 clinical isolates of P. aeruginosa. They found that 13 of these clinical isolates exhibited a strong downregulation of ErsA compared to PAO1, a finding which suggests that ErsA is under selective pressure in patients with cystic fibrosis.
Finally, Ferrara et al. established a link between ErsA and the emergence of antibiotic resistance in the treatment cystic fibrosis patients. By knocking out ErsA from a P. aeruginosa isolate (RP73) that exhibited multidrug resistance, the authors were able to reverse the resistance to some cephalosporins.
Why I selected this preprint
I selected this preLight to conclude our three-part series on respiratory tract inflammation. In the earlier preLights, Josie Gibson and I discussed the roles of inflammation and autophagy in tuberculosis. In this preprint, we see how P. aeruginosa, too, modulates the host immune system in the respiratory tract.
P. aeruginosa belongs to a group of pathogens, known as the ESKAPE pathogens, which poses multiple challenges in the arena of antibiotic resistance today. Thus far, researchers have made inroads into large molecule antimicrobials, antivirulence compounds, bacteriophages, and even host-directed therapies. By shedding new light on the role of ErsA in the pathogenicity in P. aeruginosa infections, this work by Ferrara et al. opens new opportunities to target these pathways.
Another reason for my choice to highlight this preprint is its focus on cystic fibrosis patients. Because cystic fibrosis is a rare but chronic debilitating disease, the authors’ careful study of the role of ErsA in these patients has two main benefits. The first benefit is self-evident: it pertains to the benefits that cystic fibrosis patients accrues as a direct result of this research. Better understanding the mechanisms underlying the pathogenicity of P. aeruginosa, as well as the development of antibiotic resistance, will help clinicians and patients better manage these disease complications.
The second benefit from this work relates to the applicability of the authors’ findings to other vulnerable patient populations as well. With lengthening life expectancies, a greater number of patients with multiple comorbidities is expected in the coming decades. These patients will be more vulnerable to acute infections; more patients will also suffer from chronic infections. Coupled with the looming spectre of antimicrobial resistance, better antimicrobial stewardship and management of infections will be necessary. Understanding the mediators of infection will help.
Future work
The outstanding questions from this work can be classified into three main categories. First, from an epidemiological perspective, what is the prevalence of ErsA among P. aeruginosa infections? What are the regional and global infection patterns? Is there variability across different countries or regions?
Second, how targetable is the ErsA pathway? What are some targets related to this pathway that could be potentially inhibited in order to induce bacterial cell death? Specificity is important from a pharmacological perspective, so targets that are unique to the bacteria without equivalent isoforms in mammals will be more desirable.
Third, how might modulators of the ErsA pathway interact with currently known antibiotics? The reversal of antimicrobial resistance through the knockdown of ErsA suggests that such combinations may be synergistic, but this will need to be confirmed through further studies.
These questions will not be easy to answer—it will require the collaboration of experts from different fields, among them molecular biologists, epidemiologists, microbiologists, and pharmacologists. Throughout this three-part series, we have seen how the investigation of pathogenic mechanisms underlying infectious diseases is essential to the discovery of new targets for novel antibiotics. This preprint, along with previous work conducted by the preprint authors, is a step towards that goal. I will be holding my breath.
References
[1] Miller CL, Van Laar TA, Chen T, Karna SLR, Chen P, You T, Leung KP, Global transcriptome responses including small RNAs during mixed-species interactions with methicillin-resistant Staphylococcus aureus and Pseudomonas aeruginosa, Microbiologyopen 6(3) (2017).
[2] Falcone M, Ferrara S, Rossi E, Johansen HK, Molin S, Bertoni G, The Small RNA ErsA of Pseudomonas aeruginosa Contributes to Biofilm Development and Motility through Post-transcriptional Modulation of AmrZ, Frontiers in microbiology 9 (2018) 238-238.
[3] Ferrara S, Carloni S, Fulco R, Falcone M, Macchi R, Bertoni G, Post-transcriptional regulation of the virulence-associated enzyme AlgC by the σ22-dependent small RNA ErsA of Pseudomonas aeruginosa, Environmental Microbiology 17(1) (2015) 199-214.
[4] Zhang Y-F, Han K, Chandler CE, Tjaden B, Ernst RK, Lory S, Probing the sRNA regulatory landscape of P. aeruginosa: post-transcriptional control of determinants of pathogenicity and antibiotic susceptibility, Molecular Microbiology 106(6) (2017) 919-937.
Posted on: 25 July 2020
doi: https://doi.org/10.1242/prelights.23427
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the immunology category:
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
Spatial transcriptomics elucidates medulla niche supporting germinal center response in myasthenia gravis thymoma
Prenatal inflammation reprograms hyperactive ILC2s that promote allergic lung inflammation and airway dysfunction
Also in the microbiology category:
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
Digital Microbe: A Genome-Informed Data Integration Framework for Collaborative Research on Emerging Model Organisms
Mixed Alkyl/Aryl Phosphonates Identify Metabolic Serine Hydrolases as Antimalarial Targets
Also in the pharmacology and toxicology category:
Pervasive sublethal effects of agrochemicals as contributing factors to insect decline
Optical Control of G-Actin with a Photoswitchable Latrunculin
SQ3370, the first clinical click chemistry-activated cancer therapeutic, shows safety in humans and translatability across species
preLists in the immunology category:
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |
Also in the microbiology category:
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Also in the pharmacology and toxicology category:
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
Drug use in special populations
Any drugs that are being used in special populations: Patients with liver and kidney failure, in paediatrics, in geriatrics, and in pregnant or lactating patients. Includes the discovery of factors that could potentially affect drug use in these special populations.
| List by | Zhang-He Goh |
Toxicology of toxicants, existing therapeutics, and investigational drugs
Preprints that describe the toxicology of environmental pollutants and existing and upcoming drugs. Includes both toxicokinetics and toxicodynamics, as well as technological improvements that will help in the characterisation of this field.
| List by | Zhang-He Goh |
Anticancer agents: Discovery and clinical use
Preprints that describe the discovery of anticancer agents and their clinical use. Includes both small molecules and macromolecules like biologics.
| List by | Zhang-He Goh |
Advances in Drug Delivery
Advances in formulation technology or targeted delivery methods that describe or develop the distribution of small molecules or large macromolecules to specific parts of the body.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)