The Trypanosoma brucei subpellicular microtubule array is organized into functionally discrete subdomains defined by microtubule associated proteins
Preprint posted on 9 November 2020 https://www.biorxiv.org/content/10.1101/2020.11.09.375725v1
Article now published in PLOS Pathogens at http://dx.doi.org/10.1371/journal.ppat.1009588
Categories: cell biology
Background
A key contributor to T. brucei pathogenesis is the highly asymmetric shape of its body, as it aids the parasite’s survival within the insect and mammalian vector. The trypomastigote shape of T. brucei is produced by a single layer of microtubules that underlies the plasma membrane, called the subpellicular array. A flagellum is attached to the cell surface by the flagellum attachment zone, which is comprised of a filament that is inserted between the subpellicular microtubules. Microtubules are inherently dynamic cytoskeletal polymers, and their length and activity can be altered to perform essential functions, including providing tracks for intracellular trafficking and forming the mitotic spindle. They can also be bundled to create stable structures that collectively propagate force, such as in the flagellar axoneme, which provides motility.
In T. brucei, as the flagellum beats, it deforms the subpellicular microtubule array and creates “cellular waveforms” that define the distinctive corkscrew-like motility pattern of T. brucei. The ability to regulate microtubule dynamics and flexibility within different domains of the array would allow the parasite to optimize the transmission of energy generated by the flagellar beat, and channel it into productive motility. The subpellicular array microtubules appear to be highly stable, and remain intact throughout the cell cycle, but little is known about the pathways that tune microtubule properties in trypanosomatids. In their work, Sinclair et al (1) have identified a set of T. brucei proteins that localize to different subdomains of the subpellicular array. Moreover, they characterize the location and function of the array-associated protein PAVE1, which is exclusive to kinetoplastids, and is a component of the inter-microtubule crosslinking fibrils present within the posterior subdomain. Altogether, the authors show that the subpellicular array contains various differentially localized array-associated proteins that likely function to locally tune the biophysical characteristics of the microtubules.
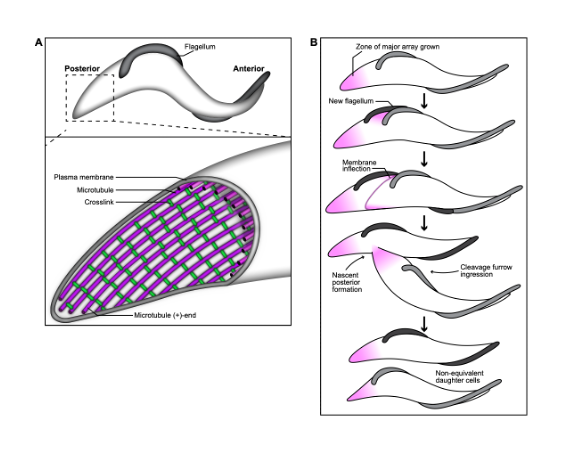
Key findings and developments
PAVE1 localizes to the inter-microtubule crosslinks of the subpellicular array at the cell posterior. PAVE1 had been previously observed to localize at the posterior and ventral edge of the subpellicular array. In this work, the authors went on to determine if PAVE1 co-localizes to regions of new microtubule growth throughout their cell cycle, by comparing PAVE1 distribution with antibody labelling of YL1/2, which recognizes the terminal tyrosine residue of alpha tubulin. Terminal tyrosination is a hallmark of newly polymerized tubulin, and a marker of array growth during the cell cycle. The authors describe PAVE1 and YL1/2 localization with respect to the number and morphology of nuclei and kinetoplasts (as markers of cell cycle). The localization pattern suggests that PAVE1 is stably associated with the array microtubules at the posterior end of the cell throughout the cell cycle, and is recruited to the nascent posterior during its formation. The authors went on to generate subpellicular array sheets from immunogold-labelled cytoskeletons, and compared PAVE1 distribution with the localization pattern of TAT1, an antibody that recognizes T. brucei a-tubulin. Comparison of TAT1 and PAVE1 showed that PAVE1 preferentially localizes to the outside ventral walls of array microtubules at the cell posterior, and to the inter-microtubule crosslinks that remained after sheet preparation. This pattern suggests that PAVE1 may be part of the inter-microtubule crosslinking fibrils present in the posterior subdomain of the array.
PAVE1 maintains microtubule length at the posterior subpellicular array. The authors went on to determine how PAVE1 and the inter-microtubule crosslinks are incorporated in the array. For this, they used HaloTag technology to visualize the localization of newly synthesized PAVE1, and how its distribution changed over time throughout 8 hours.
The authors then performed PAVE1 RNAi to test if PAVE1 is required to maintain the microtubules of the posterior array, or if array truncation in PAVE1-depleted cells is the result of aberrant cell division. They first developed a strategy to identify cells that had completed cell division prior to PAVE1 RNAi induction, and restricted the analysis to 1N2K cells to select for those that had progressed well into G1 phase at the time of PAVE1 depletion. Using an anti-tubulin antibody, the authors observed that the posterior array of PAVE1 RNAi cells at 6h and 4h was significantly shorter than at 0h. However, the length of the array between the nucleus and posterior kinetoplast was the same at all 3 time points. This suggests that PAVE1 is required to maintain the extended tapering portion of the microtubules of the cell posterior independent of its potential function during formation of a nascent cell posterior during cell division.
PAVE2 and TbAIR9 are two potential interacting partners of PAVE1. The authors investigated whether PAVE1 was part of a complex of proteins that form the inter-microtubule crosslinks at the cell posterior. To do so, they endogenously tagged both PAVE1 alleles with mNeonGreen (mNG) at the N-terminus, and immunoprecipitated mNG-PAVE1 using a mNeonTrap antibody. Two potential interacting partners were identified, namely Tb927.9.11540 a previously uncharacterized protein of 51 kDa (henceforth named PAVE2), and TbAIR9 (Tb927.11.17000, of 110 kDa). PAVE2 was located at the posterior and ventral edge of the cell which matched the localisation pattern of PAVE1, while TbAIR9 was associated with the entire array.
PAVE1 and PAVE2 require each other for stability and localization. PAVE2 was found to be conserved in kinetoplastids. PAVE2 RNAi cells were unable to divide after 4 days of RNAi induction, and showed similar division defects and posterior truncation phenotypes as PAVE1 RNAi cells. Having confirmed mirrored phenotypes upon depletion of either PAVE1 or PAVE2, the authors went on to explore what happened to localization upon depletion of either. They found that as PAVE1 levels decreased at the posterior subpellicular array, so did PAVE2, and vice versa, suggesting that PAVE1 and PAVE2 require each other for localization and protein stability inside the cell.
PAVE1 and PAVE2 form a microtubule-associated complex in vitro. As the results obtained thus far suggested that PAVE1 and PAVE2 might be part of a complex, the authors went on to test this hypothesis in vitro, and indeed suggest that PAVE1 and PAVE2 form a hetero-oligomer responsible for the construction and maintenance of the tapered cell posterior. They found also that although the PAVE complex localizes to the inter-microtubule crosslinks of the T. brucei posterior subpellicular array in vivo, they do not appear to have the capacity to crosslink microtubules in vitro. Another important observation was that the PAVE complex bound to microtubules in discrete, static, patches, which suggests that complex may recognize specific regions of microtubules. Moreover, the authors also suggest that the PAVE complex binds directly to the microtubule lattice, and may recognize a specific lattice structure, rather than a post-translational modification in the tubulin tails.
TbAIR9 controls the distribution of PAVE1 in the subpellicular array and is a global regulator of subpellicular array-associated protein localization.. Aside of PAVE2, the other potential interacting partner of PAVE1 is TbAIR9. The authors began by testing the effect of depleting TbAIR9 by RNAi on PAVE1 localisation, and found that in TbAIR9 RNAi cells, PAVE1 labelling was less intense at the cell posterior and more intense in the cell anterior compared to controls. However, PAVE1 protein levels were unaltered, indicating that TbAIR9 depletion only affects the ability of PAVE1 to localize to the posterior subdomain. Doing the mirror experiments, the authors showed that depleting PAVE1 did not affect TbAIR9 stability or localization, except for the absence at the posterior subpellicular array in cells where the posterior was truncated. With this and further investigation, the authors conclude that TbAIR9 is not necessary to build the inter-microtubule crosslinks present in the array, but it is required to properly limit the distribution of PAVE1 to the cell posterior.
The results observed upon depletion of TbAIR9 in terms of PAVE1 redistribution suggested that TbAIR9 may regulate the localization of other cytoskeletal proteins in the subpellicular array. To test this, the authors used two proteins localized at different domains of the array: Tb927.9.10790 (10790) (annotated in TrypTag as localizing to the middle of the subpellicular array), and Tb927.11.1840 (1840) (annotated in TrypTag as localized to the anterior array). Immunofluorescence imaging of both proteins on extracted cytoskeletons confirmed the expected localization and showed their stable association with the cytoskeleton. In TbAIR9 RNAi cells, both proteins lost this restricted localization, and were instead found either weakly throughout the array, or at the posterior and anterior arrays, respectively. Moreover, the protein levels of protein 10790 seemed to decrease, while levels of 1840 increased. This suggests that TbAIR9 plays a key role in organizing and confining array-associated proteins to distinct domains within the array.
What I like about this preprint
I found the work very interesting, both in terms of cell biology, and in that it advances our understanding of Trypanosoma brucei. Moreover I found the range of methods used and the range of questions addressed very varied and complete.
References
- Sinclair et al, The T. brucei subpellicular microtubule array is organized into functionally discrete subdomains defined by microtubule associated proteins, bioRxiv, 2020.
Posted on: 22 December 2020 , updated on: 2 January 2021
doi: https://doi.org/10.1242/prelights.26610
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the cell biology category:
Fetal brain response to maternal inflammation requires microglia
Alteration of long and short-term hematopoietic stem cell ratio causes myeloid-biased hematopoiesis
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
preLists in the cell biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |











 (No Ratings Yet)
(No Ratings Yet)