Two FtsZ proteins orchestrate archaeal cell division through distinct functions in ring assembly and constriction
Preprint posted on 5 June 2020 https://www.biorxiv.org/content/10.1101/2020.06.04.133736v1
Article now published in Nature Microbiology at http://dx.doi.org/10.1038/s41564-021-00894-z
A second FtsZ isoform is recruited to constrict archaea that divide without a cell wall
Selected by Dey LabCategories: biochemistry, cell biology, evolutionary biology, microbiology
Gabriel Risa [1] and Gautam Dey [1]
[1] MRC lab for Molecular Cell Biology, UCL, London WC1E 6BT, UK
Context
What are FtsZs?
FtsZs are cytoskeletal proteins belonging to the tubulin superfamily found in bacteria, archaea, and some eukaryotic organelles with bacterial ancestry, e.g. plastids (Figure 1). The superfamily is characterised by its GTPase protein domain, which is critical for the proteins’ ability to dynamically assemble into filaments and drive cell shape changes.
What is the cell biological role of FtsZ in bacteria?
FtsZ plays a key role in the bacterial cell division machinery. It is an essential protein in almost all walled bacteria and among the first to assemble at the division site, where it forms part of the Z-ring machinery responsible for cell division (Du and Lutkenhaus, 2017). During cell division, FtsZ treadmilling directs peptidoglycan septal wall synthesis that drives constriction (Bisson-Filho et al., 2017).
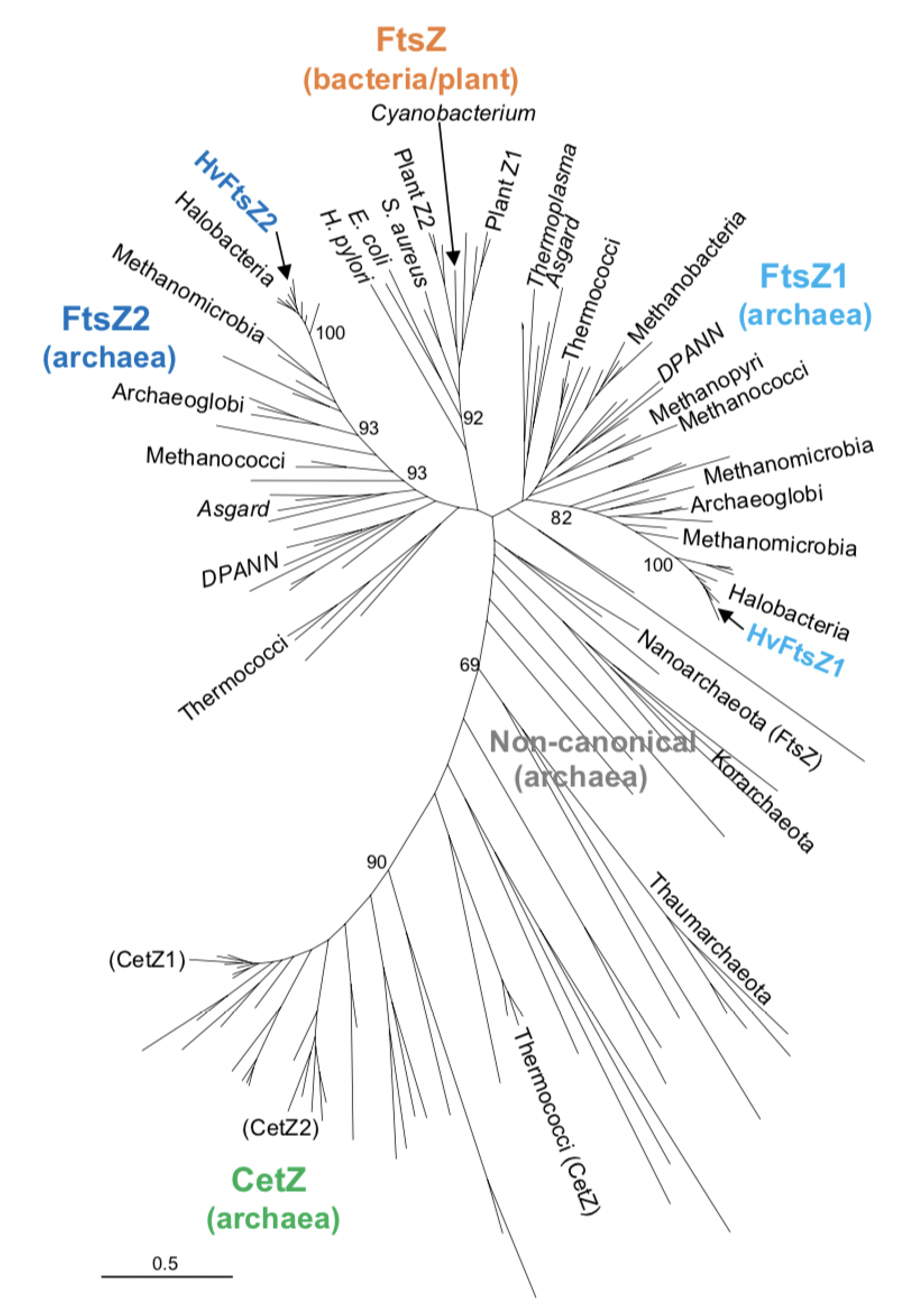
What role does FtsZs play in archaea?
FtsZs and CetZs (an archaea-specific tubulin superfamily protein) are found in several archaea (Figure 1). In contrast to bacteria, where normally only one FtsZ homologue is present, archaea often have two FtsZ homologues and numerous CetZ homologues. Intriguingly, whether an archaeon has one or two FtsZ homologues appears to coincide with whether the archaeon has a firm bacterial-like pseudomurein cell wall or a flexible S-layer, respectively.
The archaeon Haloferax volcanii has a single flexible S-layer, two FtsZ homologues, and six CetZs. None of the CetZ homologues are individually required for division, albeit at least one contributes to cell shape (Hartman et al., 2010, Duggin et al., 2015). This work by Liao, Ithurbide et al. examines the cell division of H. volcanii in the context of FtsZ. Their findings offer new insights into how two FtsZ homologues take on different roles to drive cell division in soft archaea.
Key findings
Genetic perturbations of ftsZ1 and ftsZ2 lead to division defects in H. volcanii
Remarkably and unlike bacteria, H. volcanii cells were able to grow and divide in the absence of one or both ftsZ1 and ftsZ2 genes. Nevertheless, through a series of genetic perturbations, Liao, Ithurbide et al. showed that FtsZ1 is key to cell shape and both FtsZ1 and FtsZ2 are important for the cell division process (Figure 2). Cells expressing only ftsZ1 appeared enlarged and rod-shaped, and overexpressing ftsZ1 led WT and deletion strains to form longer rod-shaped cells. In contrast, cells expressing only ftsZ2 were enlarged but discoid, and overexpressing ftsZ2 led WT cells to become smaller. In the ΔftsZ1 deletion strain, overexpressing ftsZ2 led to a partial reversion to WT morphology. In summary, it seems cells become more elongated the more FtsZ1 there is relative to FtsZ2, and overexpression of FtsZ2 can at least partially rescue the loss of FtsZ1.
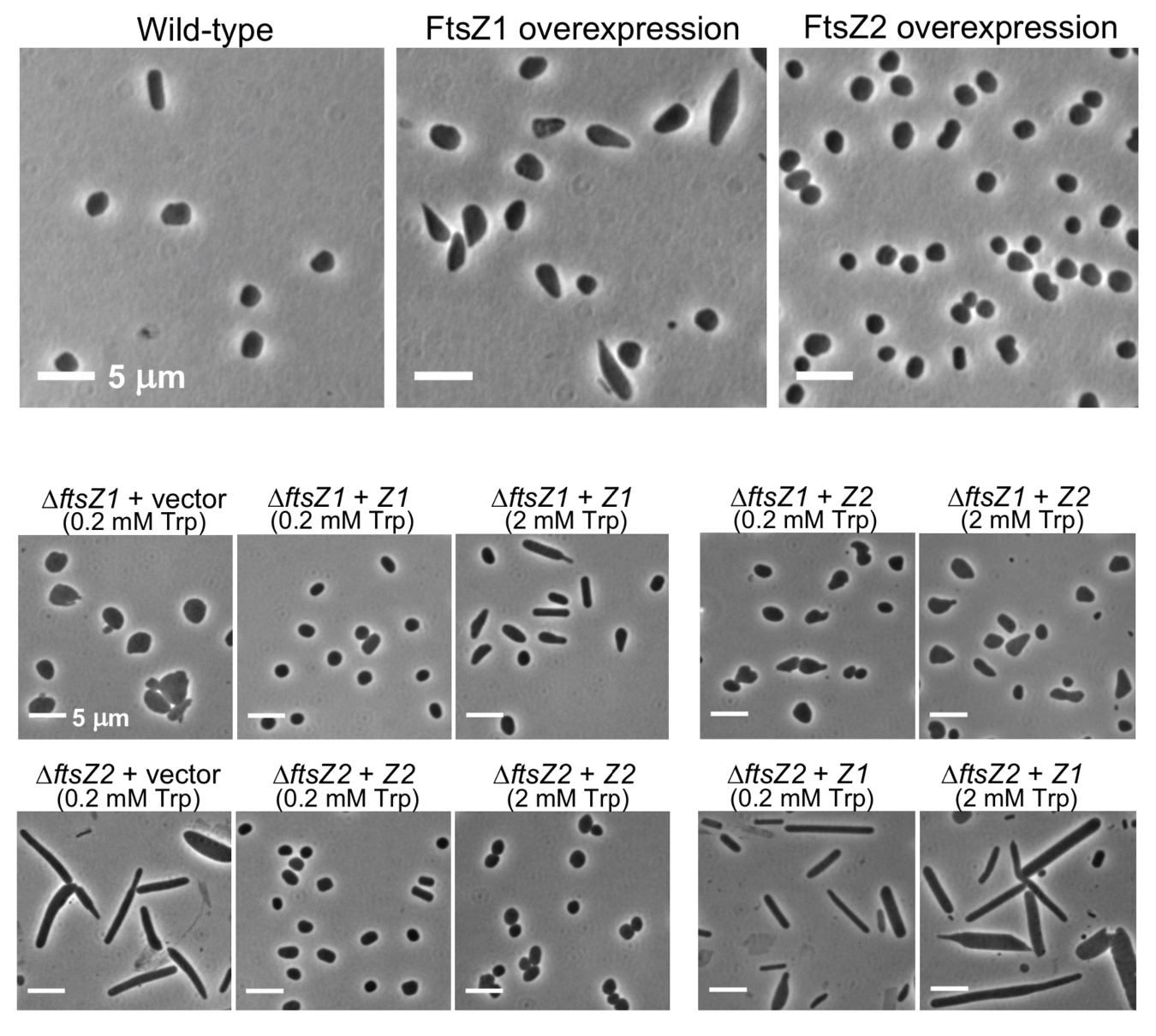
FtsZ1 and FtsZ2 collaborate during H. volcanii cell division
The authors expressed fluorescently labelled versions of ftsZ1 and ftsZ2 to observe their localisation and behaviour during cell divisions in WT and deletion strains (Figure 3). This was done at low levels of induction, as high levels of labelled proteins interfered with the division process (more a problem with FtsZ2-FP than FtsZ1-FP). FtsZ1 and FtsZ2 both co-localised at the division site and constricted together. In ΔftsZ2 cells, FtsZ1 appeared in ring-like structures, but these did not constrict. In ΔftsZ1 cells, FtsZ2 appeared in puncta and only rarely in rings. However, in some cases where FtsZ2 did appear in rings, it was also capable of constricting.
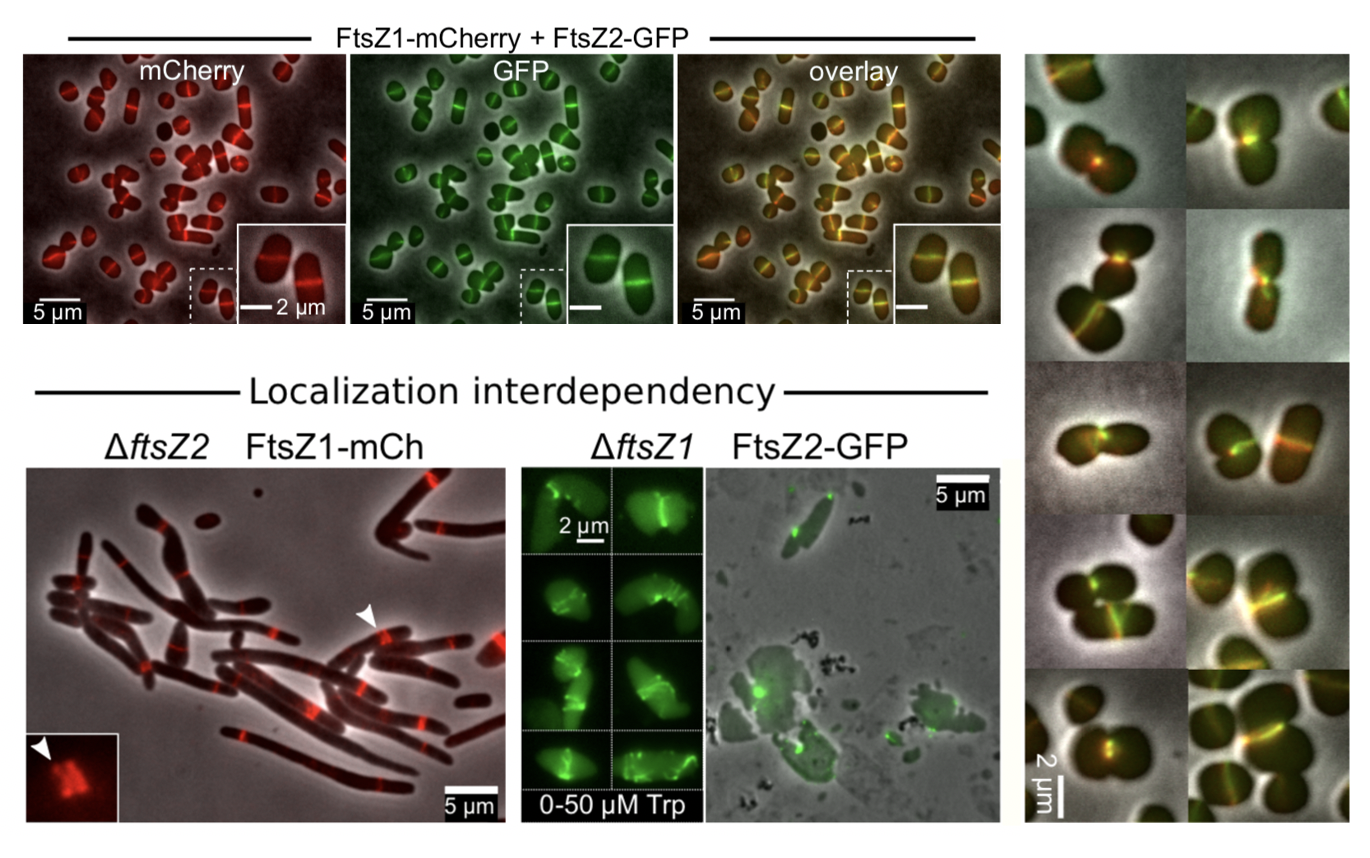
The above observations and further in-depth analysis on the degree of localisation interdependence, led the authors to propose that FtsZ1 likely plays a more structural and stabilising role compared to FtsZ2 during cell division. Furthermore, they suggest that FtsZ1 is likely recruited prior to FtsZ2 at the division site and that FtsZ2 is the homologue largely responsible for constriction.
Perspectives
Liao, Ithurbide et al. present here a working model for H. volcanii cell division. In doing so, the authors provide a suitable model organism for the study of how FtsZ mediates division in cells with a flexible S-layer, where cell wall synthesis is unlikely to drive constriction. FtsZ is one of the most conserved division proteins on the planet and therefore of great interest to the scientific community, and this work provides valuable new ground on which to build a greater understanding of its operative principles.
Questions for the authors
See questions and author response below!
References
- Bisson-Filho, A.W., Hsu, Y.P., Squyres, G.R., Kuru, E., Wu, F., Jukes, C., Sun, Y., Dekker, C., Holden, S., VanNieuwenhze, M.S., et al. (2017). Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355, 739-743.
- Du, S., and Lutkenhaus, J. (2017). Assembly and activation of the Escherichia coli divisome. Mol Microbiol 105, 177-187.
- Duggin, I.G., Aylett, C.H., Walsh, J.C., Michie, K.A., Wang, Q., Turnbull, L., Dawson, E.M., Harry, E.J., Whitchurch, C.B., Amos, L.A., et al. (2015). CetZ tubulin-like proteins control archaeal cell shape. Nature 519, 362-365.
- Hartman, A.L., Norais, C., Badger, J.H., Delmas, S., Haldenby, S., Madupu, R., Robinson, J., Khouri, H., Ren, Q., Lowe, T.M., et al. (2010). The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PLoS One 5, e9605.
Posted on: 25 June 2020
doi: https://doi.org/10.1242/prelights.22237
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the biochemistry category:
Structural basis of respiratory complexes adaptation to cold temperatures
Lens Placode Modulates Extracellular Matrix Formation During Early Eye Development
Generalized Biomolecular Modeling and Design with RoseTTAFold All-Atom
Also in the cell biology category:
Clusters of lineage-specific genes are anchored by ZNF274 in repressive perinucleolar compartments
Structural basis of respiratory complexes adaptation to cold temperatures
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
Also in the evolutionary biology category:
How the liver contributes to stomach warming in the endothermic white shark Carcharodon carcharias
A long non-coding RNA at the cortex locus controls adaptive colouration in butterflies
AND
The ivory lncRNA regulates seasonal color patterns in buckeye butterflies
AND
A micro-RNA drives a 100-million-year adaptive evolution of melanic patterns in butterflies and moths
A complex Plasmodium falciparum cryptotype circulating at low frequency across the African continent
Also in the microbiology category:
RIPK3 coordinates RHIM domain-dependent inflammatory transcription in neurons
Digital Microbe: A Genome-Informed Data Integration Framework for Collaborative Research on Emerging Model Organisms
Mixed Alkyl/Aryl Phosphonates Identify Metabolic Serine Hydrolases as Antimalarial Targets
preLists in the biochemistry category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
Preprint Peer Review – Biochemistry Course at UFRJ, Brazil
Communication of scientific knowledge has changed dramatically in recent decades and the public perception of scientific discoveries depends on the peer review process of articles published in scientific journals. Preprints are key vehicles for the dissemination of scientific discoveries, but they are still not properly recognized by the scientific community since peer review is very limited. On the other hand, peer review is very heterogeneous and a fundamental aspect to improve it is to train young scientists on how to think critically and how to evaluate scientific knowledge in a professional way. Thus, this course aims to: i) train students on how to perform peer review of scientific manuscripts in a professional manner; ii) develop students' critical thinking; iii) contribute to the appreciation of preprints as important vehicles for the dissemination of scientific knowledge without restrictions; iv) contribute to the development of students' curricula, as their opinions will be published and indexed on the preLights platform. The evaluations will be based on qualitative analyses of the oral presentations of preprints in the field of biochemistry deposited in the bioRxiv server, of the critical reports written by the students, as well as of the participation of the students during the preprints discussions.
| List by | Marcus Oliveira |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
20th “Genetics Workshops in Hungary”, Szeged (25th, September)
In this annual conference, Hungarian geneticists, biochemists and biotechnologists presented their works. Link: http://group.szbk.u-szeged.hu/minikonf/archive/prg2021.pdf
| List by | Nándor Lipták |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
MitoList
This list of preprints is focused on work expanding our knowledge on mitochondria in any organism, tissue or cell type, from the normal biology to the pathology.
| List by | Sandra Franco Iborra |
Also in the cell biology category:
BSCB-Biochemical Society 2024 Cell Migration meeting
This preList features preprints that were discussed and presented during the BSCB-Biochemical Society 2024 Cell Migration meeting in Birmingham, UK in April 2024. Kindly put together by Sara Morais da Silva, Reviews Editor at Journal of Cell Science.
| List by | Reinier Prosee |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
CellBio 2022 – An ASCB/EMBO Meeting
This preLists features preprints that were discussed and presented during the CellBio 2022 meeting in Washington, DC in December 2022.
| List by | Nadja Hümpfer et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
FENS 2020
A collection of preprints presented during the virtual meeting of the Federation of European Neuroscience Societies (FENS) in 2020
| List by | Ana Dorrego-Rivas |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
Cell Polarity
Recent research from the field of cell polarity is summarized in this list of preprints. It comprises of studies focusing on various forms of cell polarity ranging from epithelial polarity, planar cell polarity to front-to-rear polarity.
| List by | Yamini Ravichandran |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
3D Gastruloids
A curated list of preprints related to Gastruloids (in vitro models of early development obtained by 3D aggregation of embryonic cells). Updated until July 2021.
| List by | Paul Gerald L. Sanchez and Stefano Vianello |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
ASCB EMBO Annual Meeting 2019
A collection of preprints presented at the 2019 ASCB EMBO Meeting in Washington, DC (December 7-11)
| List by | Madhuja Samaddar et al. |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Lung Disease and Regeneration
This preprint list compiles highlights from the field of lung biology.
| List by | Rob Hynds |
Cellular metabolism
A curated list of preprints related to cellular metabolism at Biorxiv by Pablo Ranea Robles from the Prelights community. Special interest on lipid metabolism, peroxisomes and mitochondria.
| List by | Pablo Ranea Robles |
BSCB/BSDB Annual Meeting 2019
Preprints presented at the BSCB/BSDB Annual Meeting 2019
| List by | Dey Lab |
Biophysical Society Annual Meeting 2019
Few of the preprints that were discussed in the recent BPS annual meeting at Baltimore, USA
| List by | Joseph Jose Thottacherry |
ASCB/EMBO Annual Meeting 2018
This list relates to preprints that were discussed at the recent ASCB conference.
| List by | Dey Lab, Amanda Haage |
Also in the evolutionary biology category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
preLights peer support – preprints of interest
This is a preprint repository to organise the preprints and preLights covered through the 'preLights peer support' initiative.
| List by | preLights peer support |
EMBO | EMBL Symposium: The organism and its environment
This preList contains preprints discussed during the 'EMBO | EMBL Symposium: The organism and its environment', organised at EMBL Heidelberg, Germany (May 2023).
| List by | Girish Kale |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
EMBL Synthetic Morphogenesis: From Gene Circuits to Tissue Architecture (2021)
A list of preprints mentioned at the #EESmorphoG virtual meeting in 2021.
| List by | Alex Eve |
Planar Cell Polarity – PCP
This preList contains preprints about the latest findings on Planar Cell Polarity (PCP) in various model organisms at the molecular, cellular and tissue levels.
| List by | Ana Dorrego-Rivas |
TAGC 2020
Preprints recently presented at the virtual Allied Genetics Conference, April 22-26, 2020. #TAGC20
| List by | Maiko Kitaoka et al. |
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
COVID-19 / SARS-CoV-2 preprints
List of important preprints dealing with the ongoing coronavirus outbreak. See http://covidpreprints.com for additional resources and timeline, and https://connect.biorxiv.org/relate/content/181 for full list of bioRxiv and medRxiv preprints on this topic
| List by | Dey Lab, Zhang-He Goh |
1
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |
Pattern formation during development
The aim of this preList is to integrate results about the mechanisms that govern patterning during development, from genes implicated in the processes to theoritical models of pattern formation in nature.
| List by | Alexa Sadier |
Also in the microbiology category:
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)