Upregulation and cell specificity of C4 genes are derived from ancestral C3 gene regulatory networks
Posted on: 30 July 2020
Preprint posted on 4 July 2020
Categories: plant biology
Background:
How do complex traits evolve and what is the genetic basis for complex traits? This has been a basic question that has motivated many researchers to study how the rewiring of gene regulatory networks alters downstream physiological responses of many organisms (Sun and Dinneny 2018). In plant biology, a complex trait that has been used as a model to understand the evolution of adaptive traits has been the transition of plants that perform C3 photosynthesis to C4 photosynthesis (Osborne and Sack 2012). All plants perform photosynthesis using the enzyme Rubisco to turn light, carbon dioxide and water into sugars that fuel plant growth. While the majority of plant species on earth uses C3 photosynthesis, during this process, Rubiso inefficiently interacts with O2 instead of CO2 to create a toxic compound that requires recycling through photorespiration (Singh et al. 2020). Over time, plants have evolved methods to compartmentalize photosynthesis to reduce this inefficiency and improve water retention. C4 photosynthesis is a complex trait characterized by changes in anatomy, biochemistry, and gene expression. Instead of conducting C3 photosynthesis in the mesophyll cells, C4 photosynthesis delivers CO2 to Rubisco in bundle sheath cells (Aubry et al. 2014). The convergent evolution of C4 photosynthesis from C3 ancestors, which include 62 independent lineages accounting for ~8,100 species, makes this network an attractive area to study how the gene networks underpinning C4 photosynthesis are derived from those that operate in C3 ancestors (Singh et al. 2020).
Key findings:
In this preprint, Singh et al., are interested in defining the transcription factors and gene expression dynamics associated with C4 photosynthesis in the dicot Gynandropsis gynandra. They first defined the induction of C4 photosynthesis by observing how seedlings grown in the dark will respond once transferred to light. They monitored the dynamics of photosynthesis induction to define timepoints to profile the transcript abundance changed during C4 photosynthesis in Gynandropsis gynandra and compared this with C3 photosynthesis in the model plant, Arabidopsis.

Figure 1: Gynandropsis gynandra seedlings greening and unhooking cotyledons.
They identified transcription factors associated with C4 photosynthesis and found that while many genes associated with core photosynthetic processes are shared by C3 and C4 plants during dark to light transition, transcript abundance is higher in the C4 plant. Using DNase-Seq to explore chromatin accessibility and transcription factor footprints along the genome, the authors identified differences in transcription factor binding sites to be a large contributor in activating C4 photosynthesis genes as opposed to open chromatin dynamics. By comparing the cistrome of C3 and C4 photosynthesis genes from G. gynandra and the C3 photosynthesis genes from Arabidopsis, the group proposed two models related to the molecular evolution of C4 photosynthesis. 1. Increased expression of C4 genes are associated with the increased binding frequency of transcription factors to EE-Box, I-Box, and C2C2-GATA binding sites. 2. Increased transcription factor binding to homeodomain binding sites may be associated with mesophyll specific expression. These findings contribute toward our understanding of how complex traits evolve over time and the necessary components required to install C4 traits into C3 plants, thereby potentially improving the photosynthetic efficiency of our crop plants.
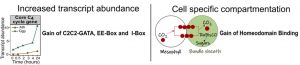
Figure 2: Model illustrating gain of cis-elements bound by MYB-related and C2C2-GATA transcription factors as well as the gain of homeodomain binding sites in mesophyll expressed genes in C4 G. gynandra.
Questions to the authors?
Q1: The authors here have focused on chromatin dynamics and transcriptional changes occurring before 2 hours of light treatment. I wonder what is happening to the gene network after 2 hours that is causing the second phase of photosynthetic induction.
Q2: What are the limitations and next steps toward identifying the transcription factors that are key targets for engineering the C4 pathway into C3 crops.
Q3: How were LREs and homeodomains gained or co-opted during C4 evolution and are these motifs also over-represented in other species that have independently evolved C4 photosynthesis?
Further readings and References
Aubry, Sylvain, Steven Kelly, Britta M. C. Kümpers, Richard D. Smith-Unna, and Julian M. Hibberd. 2014. “Deep Evolutionary Comparison of Gene Expression Identifies Parallel Recruitment of Trans-Factors in Two Independent Origins of C4 Photosynthesis.” PLoS Genetics 10 (6): e1004365.
Burgess, Steven J., Ivan Reyna-Llorens, Sean R. Stevenson, Pallavi Singh, Katja Jaeger, and Julian M. Hibberd. 2019. “Genome-Wide Transcription Factor Binding in Leaves from C3 and C4 Grasses.” The Plant Cell 31 (10): 2297–2314.
Osborne, Colin P., and Lawren Sack. 2012. “Evolution of C4 Plants: A New Hypothesis for an Interaction of CO2 and Water Relations Mediated by Plant Hydraulics.” Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 367 (1588): 583–600.
Singh, P., S. R. Stevenson, I. Reyna-Llorens, and G. Reeves. 2020. “Upregulation and Cell Specificity of C4 Genes Are Derived from Ancestral C3 Gene Regulatory Networks.” bioRxiv. https://www.biorxiv.org/content/10.1101/2020.07.03.186395v1.abstract.
Sun, Ying, and José R. Dinneny. 2018. “Q&A: How Do Gene Regulatory Networks Control Environmental Responses in Plants?” BMC Biology 16 (1): 38.
doi: https://doi.org/10.1242/prelights.23553
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the plant biology category:
Actin Counters Geometry to Guide Plant Cell Division
Jeny Jose
The nucleus follows an internal cellular scale during polarized root hair cell development
Jeny Jose
Conservation and divergence of regulatory architecture in nitrate-responsive plant gene circuits
Jeny Jose
preLists in the plant biology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
The Society for Developmental Biology 82nd Annual Meeting
This preList is made up of the preprints discussed during the Society for Developmental Biology 82nd Annual Meeting that took place in Chicago in July 2023.
| List by | Joyce Yu, Katherine Brown |
CSHL 87th Symposium: Stem Cells
Preprints mentioned by speakers at the #CSHLsymp23
| List by | Alex Eve |
SDB 78th Annual Meeting 2019
A curation of the preprints presented at the SDB meeting in Boston, July 26-30 2019. The preList will be updated throughout the duration of the meeting.
| List by | Alex Eve |











 (No Ratings Yet)
(No Ratings Yet)