preLights talks to David Stephens
19 July 2021
David Stephens is Professor of Cell Biology in the School of Biochemistry at the University of Bristol. His research focuses on the cell biology of membranes and cytoskeletal dynamics, which his lab investigates using advanced light microscopy techniques, 3D cell culture and zebrafish. He is an affiliate for bioRxiv and is also an editor for the Journal of Cell Science, published by The Company of Biologists. We spoke to David about his research career, the importance of a positive research community and why he advocates for preprinting.

To start – what inspired your interest in biochemistry and what led to you pursuing that as a degree?
I’d always been interested in biology, so I knew I wanted to do something biology related. It was made more obvious to me at school that biochemistry gave me more career options than botany or zoology, which were still quite common at the time. There were a few broader biology degrees but they didn’t grab me quite so much: once we started learning about DNA replication and protein synthesis I realised that was pretty cool, so I really chose it because it was what I was most interested in. To be honest, I think everyone should choose a degree programme because they’re interested in it!
Then you pursued a PhD with Brian Austen at St George’s, followed by a postdoc with George Banting at the University of Bristol. Can you tell us about that time?
My PhD was really my first foray into cell biology and trafficking, looking into the formation of the amyloid peptide and how this related to the trafficking itinerary of the precursor protein. For my postdoc, I moved onto an endocytosis project looking at the role of clathrin adaptors, and I had a real interest in membrane dynamics and how things moved around cells. That was a really good time in the field because there was an awful lot of new insight coming from structural biology as well as biochemistry and cell biology. There was a great group of people working in the lab and contributing to projects in different ways, and my time as a postdoc there gave me proper training in scientific methods and the route of taking a project from conception to completion.
This led to a second postdoc with Rainer Pepperkok at the European Molecular Biology Laboratory (EMBL) in Heidelberg, focusing on COPI and COPII, which you’ve maintained a long-term interest in.
I’d only worked on clathrin adaptors with George, so the move to EMBL meant a shift to a slightly different vesicle trafficking system. This was a less mature field; a lot had been answered in the field of endocytosis, but what we knew about COPI and COPII was still anchored in in vitro systems, and we didn’t know very much at all about how this operated in mammalian cells. This seemed like a great combination of a problem waiting to be looked at, with the solution, which was using live-cell imaging, and Rainer’s lab was a great place to go to for that. As we’ve got deeper into the subject we’ve realised that there’s an awful lot more to it than we originally thought, and that’s really sustained my interest. The complexity is one of the most interesting things about this field: one-third of the human genomes encodes proteins that traffic through the secretory pathway, and when you start looking at different cell types or cells in different contexts, there’s still so much to be looked at.
This is also a really nice field to be in. It is super competitive, but it’s been great throughout. It doesn’t matter whether you’re a young postdoc starting out, or someone who has been in the field for a while, the people are nice and they’ll come and speak with you at conferences. It’s been great to engage with some of the superstars of the field; they care about the science, and they’ll be generous with their time and advice. I think that’s the kind of thing that sustains you in a field, and I’ve found a similar community more recently with moving into cilia-based work.
Now you’re well known for cell imaging – did you approach these techniques because you were specifically interested in them, or was this driven by your research questions?
I wanted to learn these techniques primarily to answer biological questions. I’ve never had any formal physics or optics training that allows me to build my own systems, so whenever we make tentative steps in that sort of direction it’s always in collaboration with experts in the field. EMBL is often used as a testing site for companies to bring in their latest equipment, so that was a great opportunity to have a play with new kit. I think it’s important to find the right technique that answers your biological question, and it’s nice to see that field continue so actively with the latest developments in 3D electron microscopy and super-resolution imaging, which opens up new avenues for answering questions as well.
I do think we need constant vigilance to make sure there’s continued awareness of the skills of technical staff and the importance of core facilities infrastructure, and that this should be reflected in funding policies.
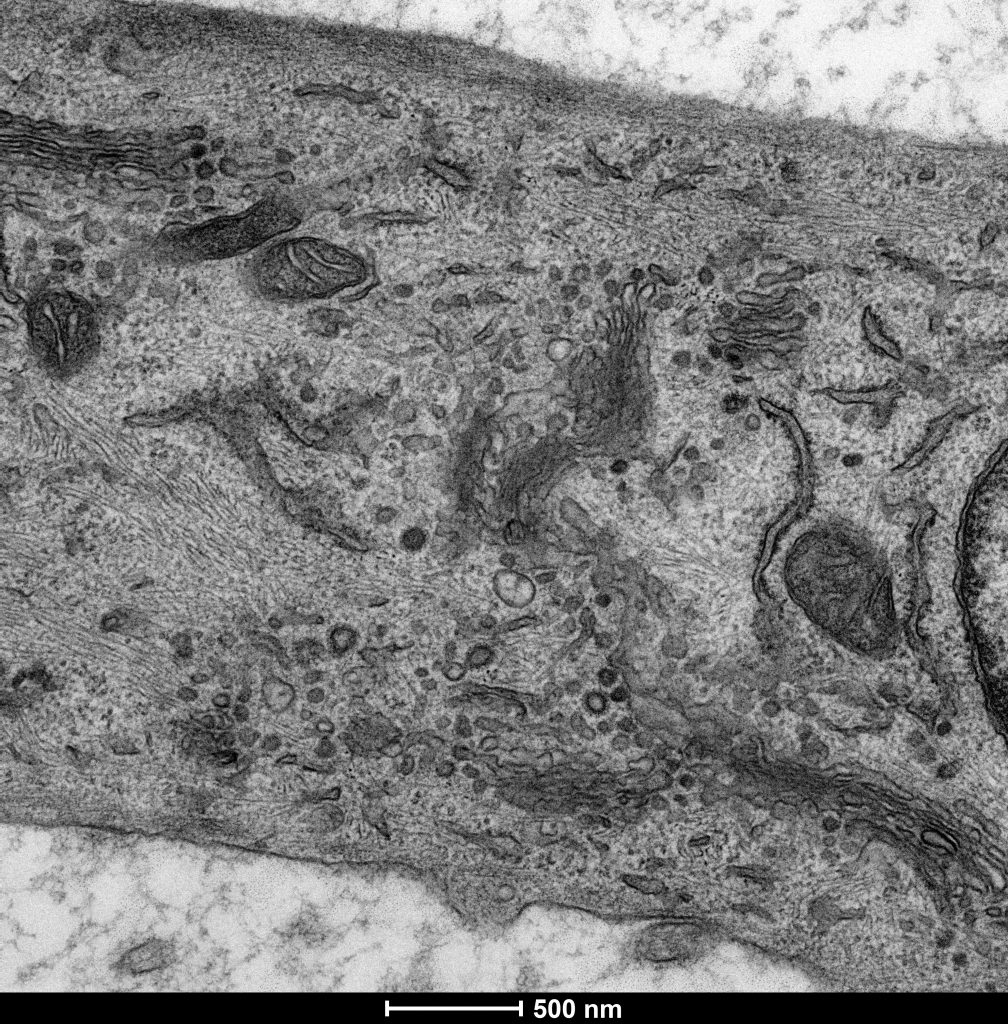
Electron micrograph of a mammalian cell (Stephens lab)
As you mentioned, you’ve started to focus on ciliogenesis and the role of the dynein-2 complex. What led to that new pathway in your research?
When I set up my own lab in Bristol, the idea was to build on what I’d started working on with Rainer about integrating microtubule function with early secretory pathway functions. We looked at the roles of dynein and dynactin that match the classical cytoplasmic dynein, which is dynein-1, and we started doing some work involving RNA depletion of all the individual dynein subunits in the cell. I thought we needed a really good control, and when I started looking in the literature I realised there was a dynein-2 that might also have something to do with Golgi function as well. But there was very little known about the dynein-2 motor, essentially, in terms of mammalian cells – just that there was a heavy chain and a light intermediate chain. So it led to us getting really interested in this motor and using it as a negative control of trafficking, which meant that our positive control was ciliogenesis! The more we looked at cilia in the lab the more the work just took off and became something I wanted to keep doing. I was happy to broaden what we do as a lab and keep aware of more diverse views on the way that membranes and cytoskeletal components interact.
Have you always thought about how your research can be applied in new contexts?
We always think about the more applied possibilities and why people would want to know about what we’re doing. As you become known in a field and more established as a researcher you’re able to contact people and start working on different systems and approaching new systems.
On my website I talk about this ‘nano to Danio’ approach, which came about because a lot of the mutations to early secretory pathways genes result in abnormal skeletal formation, including craniofacial defects. Zebrafish are very tractable for things like that because they have a bony skeleton. All of our zebrafish work is in collaboration with Chrissy Hammond, and although we might not get as far as we would like with physiological relevance, we do get some really nice insight into tissue function. For developmental biology and the broader extracellular matrix concept, we’re part of a much larger team led by Karl Kadler and involving a lot of people at the University of Manchester. The same for imaging – dabbling in using higher-resolution imaging models has been in collaboration with physicists like Andrew Hudson, who’s now at Leicester. Working with him has helped us get as much as we can out of our data without having to spend three years on a project to develop a new tool.
These collaborations really give us more context for our work, and it’s much more pleasurable to be able to do this team-based science. You can’t be an expert in everything.
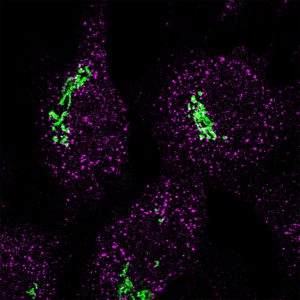
Immunofluorescence image showing Sec16a (magenta) and GRASP65 (green) (Stephens lab)
You did a postdoc at Bristol University and subsequently established your own lab there too. Were you particularly drawn to the biochemistry department there?
It’s a lovely place to be – scientifically, academically, all of those things, but it’s also a great place to live. That matters to me, and I don’t think enough people necessarily give it enough consideration when choosing a job. I knew George very well by that point and there were other people there, including Steve Halford, who were very influential in making that decision too. I’d got to know Steve well and was friends with people who used to work in his lab who had also set up their own labs at Bristol, so I was positively encouraged to come back and carve out my own niche. There was also a well-established imaging facility, so it became a logical place to go.
And at Bristol you’ve been quite involved in undergraduate teaching. In fact, as organiser for the fourth year of the MSci Biochemistry degree, the course reported 100% student satisfaction! Is engaging with undergraduates as well as PhD students important to you?
The emergence of these four-year MSci degrees is a great aspect of the UK university system, and it was a really nice programme to be involved with setting up because it helps embed that research ethos into undergraduate training. It’s really nice to see students take their first steps into independent research, critical analysis and mapping out data. In many ways, I think that fourth year is relatively similar to the first year of a PhD.
Recently, I’ve taken a bit of a step back from teaching as I’m doing more administrative roles, but I fully intend to come back to it and do more of it. It’s really nice to see that journey through the university and feel that you can make a positive difference to what they’re doing. I think there’s a perception that we expect all undergraduates to do PhDs and become researchers, but I’m always happy when students want to go and do jobs in the ‘regular’ world, too. Doing a biochemistry degree does not define you as a biochemist for the rest of your career, and I’ll try to point students to the best people who can give them the right advice.
Moving onto preprinting. When did you become aware of this and were you quick to adopt it?
I think I’d always been aware of aRxiv in the background for the physics field, but I’d never really thought about it in terms of biosciences. Richard Sever, of Cold Spring Harbor Laboratory, got in touch with me before bioRxiv launched and signed me up as an affiliate, and I remember discussing the plans and thinking it was an interesting concept; I couldn’t really think of why you wouldn’t want to do it to be honest! Of course, since then, people have come up with various reasons as to why they don’t want to preprint their work – so I think it’s quite an individual decision and also quite field specific.
I was a relatively early adopter of preprints, and now it’s pretty much lab policy to preprint, and we will always try to do it. I’m fully behind it, and it would be my intention to preprint every piece of work that we publish. I suppose being an affiliate since 2013 has been a driving part of that narrative, but my caveat is that I wouldn’t force anyone in my lab to do it.
For early-career researchers, what do you think the main advantages of preprinting their work are?
Mainly, it’s about sharing your work, and showing the accomplishments that you’ve made in your research without waiting for the peer-review publication cycle that can take so long. When you’re going for jobs or applying for fellowships, there’s a record of output, even if it’s at a slightly earlier stage than a fully peer-reviewed publication. You can still see how you’ve set out your ideas in the text, your figures, your controls, and I think having the ability to showcase your work is the biggest positive by far.
I’ve used preprinting very widely in all aspects of my job, including using it to secure a grant. We saw a preprint come out last year as were finalising a grant application and immediately got in touch with the lead author to set up a collaboration, which went straight into the grant application. Preprints work!
How do you think preLights plays a role in the preprint landscape?
There are an awful lot of preprints published every single day, and nobody’s going to be able to read them all. Having some insight into which ones are good to look at, especially from early-career researchers is great, because it might be for different reasons than someone like me would choose. It’s a very positive process to choose a preprint to highlight.
And to finish, what one piece of advice would you give to early-career researcher who would like to pursue a long-term career in academia?
I’d actually like to reframe this question to one about any early-career researcher thinking about pursuing a long-term career in research. That doesn’t have to be in academia. There are so many jobs in science outside of academia that still contribute to the research effort – you look at somewhere like Bristol, we’ve got so many spin-out science companies in the vicinity of us, and there are so many opportunities associated with innovation and enterprise. So, I think that’s my major bit of advice: don’t view it as a career in academia, consider it a career in research more widely.
David was interviewed by Helen Robertson, Community Manager for preLights. This interview has been approved by the interviewee.










