DeepImageTranslator: a free, user-friendly graphical interface for image translation using deep-learning and its applications in 3D CT image analysis
Posted on: 3 June 2021
Preprint posted on 17 May 2021
Article now published in SLAS Technology at http://dx.doi.org/10.1016/j.slast.2021.10.014
DeepImageTranslator: A new open-access graphical interface that enables researchers with no programming experience to design and evaluate deep-learning models for image translation applications.
Selected by Afonso MendesCategories: bioinformatics
Background:
Imaging is a fundamental tool in biomedical research, but its use requires the interpretation of the images acquired to detect and annotate relevant features (i.e., image classification). The development of imaging frameworks led to the establishment of high- throughput methods that generate large image libraries. Thus, manually processing such large-scale datasets becomes impractical, and prone to a high degree of error and low reproducibility. Deep-learning algorithms are computer processing systems with an architecture inspired by the neural networks composing animal brains. Artificial neural networks, usually called convolutional neural networks (CNNs), consist of interconnected layers of nodes (or neurons), where each neuron receives input from the neurons in the previous layer and sends a processed output to all the neurons in the next layer. CNNs excel at performing tasks involving pattern detection and decision-making. The advent of deep-learning algorithms had a disruptive effect in image translation, drastically improving the ability to analyse large image datasets and extract relevant information[1-3]. However, using these methods typically requires computer programming experience, which limits their application in biomedical research. A small number of open-access graphical interfaces enabling researchers with no programming experience to apply deep-learning algorithms to image translation are available, but they usually deploy previously designed algorithms with a fixed architecture[4]. In this preprint, Zhou et al. developed a simple and free graphical interface that allows inexperienced users to create, train, and evaluate their own deep-learning models for image translation.
Key Findings:
1) Development of DeepImageTranslator: a simple and open-access graphical interface that enables the creation, training, and evaluation of deep-learning pipelines for image translation.
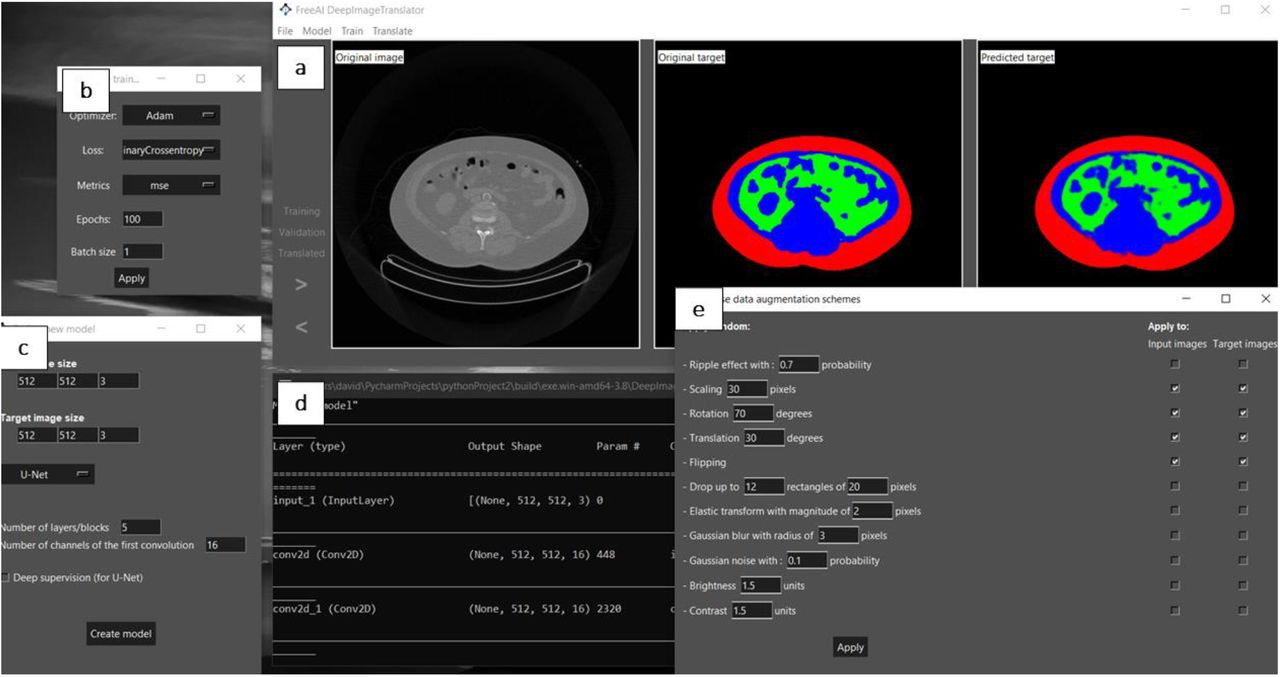
The authors start by presenting the software developed. It is a simple and open- access graphical interface encompassing key features. First, it includes a main window to visualize the training, validation, and test datasets (Fig. 1a). Another window contains options to select the type of model optimiser, loss function, training metrics, batch size, and number of epochs/iterations (Fig. 1b). Moreover, it includes a window that enables the modulation of the CNN’s architectural features, such as the number and type of convolutional layers (Fig. 1c). It also includes a window to monitor the training process (Fig. 1d) and another with options to modulate the data augmentation scheme (Fig. 1e). The neural network employed follows the general structure of U-net[5].
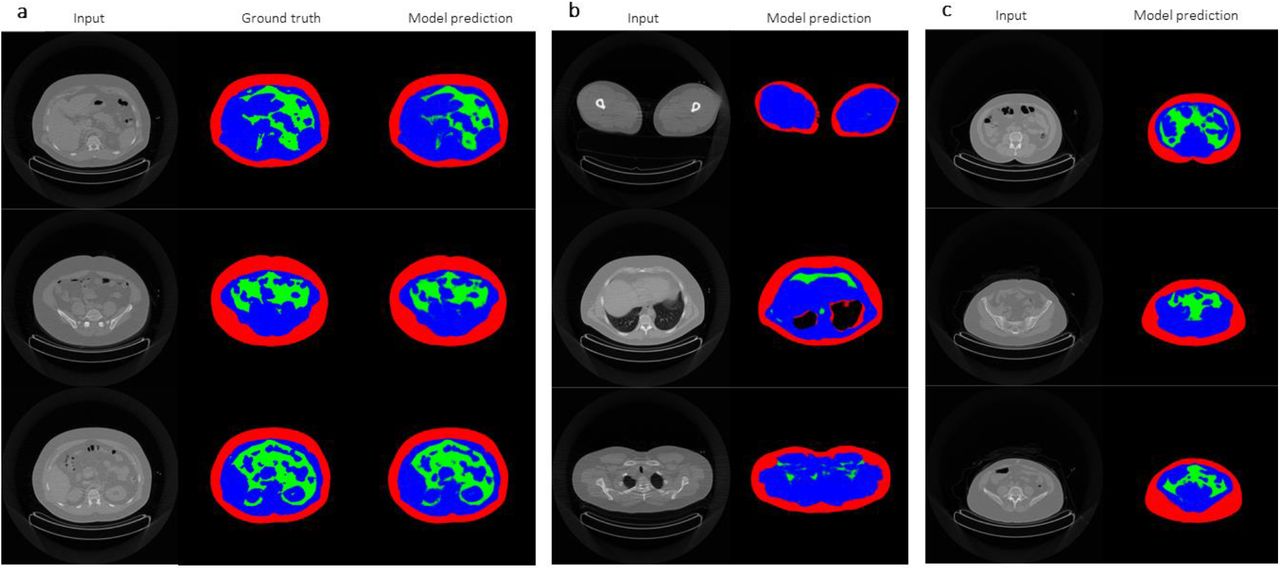
2) DeepImageTranslator can be applied to computed tomography (CT) datasets for segmentation tasks and produces models with a high degree of generalisability.
In the next sections, the authors demonstrate the application of their software using CT image libraries. The segmentation of different features in an image is a crucial task in image translation. The authors create a CNN capable of performing a segmentation task involving the differentiation of subcutaneous adipose tissue, visceral adipose tissue, and lean tissues from CT images (Fig. 2a). The generalisability of the model is showcased in several manners. The neural network was capable of performing the segmentation task on CT images of legs, thorax, and scapular regions, even though it was trained using images from the abdominal region (Fig. 2b). Moreover, the model performed well regardless of the subjects’ bodyweight, body composition, and gender (Fig. 2c). Importantly, the model was able to achieve a remarkably high predictive power using a sample as small as 17 images, outperforming previously reported models based on significantly larger datasets.
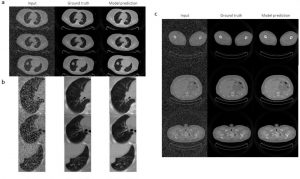
3) DeepImageTranslator enables noise reduction for thoracic CT images.
The versatility of the neural networks produced using DeepImageTranslator is finally showcased by using the software for another crucial task in image translation – noise reduction. The authors used a dataset containing CT images of thoracic regions further manipulated to become noisy. The model was able to considerably reduce the noise introduced in the images and produce predictions that were almost indistinguishable from the noiseless images (Fig. 3). Interestingly, the model was able to recover details of the pulmonary vasculature invisible on the noisy images.
Why I think this work is important:
As stated throughout this post, deep-learning models excel in performing crucial image translation tasks for biomedical research, such as segmentation and noise reduction. A major caveat for the application of this approach is that it usually requires programming experience, which is not a common skill among biomedical researchers. The development of tools such as DeepImageTranslator facilitate the access of inexperienced users to this technology and is important to enable its widespread application. While other projects that provide user-friendly access to deep-learning tools for image translation are available[4], DeepImageTranslator goes one step further and enables the user to modulate architectural elements of the neural network and evaluate its capability to perform specific tasks.
Questions for the authors:
- Do you plan to support the software, for example, by providing updates in the future? If so, did you consider including a framework to share deep-learning pipelines between users?
- Although you demonstrate that commonly used computers were able to perform the intended tasks in 5 to 17 hours, do you plan to use cloud-based processing to enable users with low-end computers to employ your software?
References:
[1] Yasaka et al. (2018) Deep learning with convolutional neural network in radiology. Jpn J Radiol. 36(4):257-272. Doi: 1007/s11604-018-0726-3.
[2] Chi et al. (2019) Computed Tomography image quality enhancement via a uniform framework integrating noise estimation and super-resolution networks. Sensors. 19(15):3348. Doi: 10.3390/s19153348.
[3] Xiang et al. (2018) Deep embedding convolutional neural network for synthesizing CT image from T1-Weighted MR image. Medical Image Analysis. 47:31-44. Doi: 10.1016/j.media.2018.03.011.
[4] Chamier et al. (2021) Democratising deep learning for microscopy with ZeroCostDL4Mic. Nature Communications. 12:2276. Doi: 1038/s41467-021-22518-0.
[5] Falk et al. (2019) U-Net: deep learning for cell counting, detection, and morphometry. Nature Methods. 16:67-70. Doi: 1038/s41592-018-0261-2.
doi: https://doi.org/10.1242/prelights.29327
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioinformatics category:
The lipidomic architecture of the mouse brain
CRM UoE Journal Club et al.
Kosmos: An AI Scientist for Autonomous Discovery
Roberto Amadio et al.
Human single-cell atlas analysis reveals heterogeneous endothelial signaling
Charis Qi
preLists in the bioinformatics category:
Keystone Symposium – Metabolic and Nutritional Control of Development and Cell Fate
This preList contains preprints discussed during the Metabolic and Nutritional Control of Development and Cell Fate Keystone Symposia. This conference was organized by Lydia Finley and Ralph J. DeBerardinis and held in the Wylie Center and Tupper Manor at Endicott College, Beverly, MA, United States from May 7th to 9th 2025. This meeting marked the first in-person gathering of leading researchers exploring how metabolism influences development, including processes like cell fate, tissue patterning, and organ function, through nutrient availability and metabolic regulation. By integrating modern metabolic tools with genetic and epidemiological insights across model organisms, this event highlighted key mechanisms and identified open questions to advance the emerging field of developmental metabolism.
| List by | Virginia Savy, Martin Estermann |
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |











 (No Ratings Yet)
(No Ratings Yet)