Functional Diversity of Memory CD8 T Cells is Spatiotemporally Imprinted
Posted on: 22 August 2024 , updated on: 5 March 2025
Preprint posted on 25 March 2024
Article now published in Nature at http://dx.doi.org/10.1038/s41586-024-08466-x
Know your neighbors – spatial positioning determines tissue-resident memory T cell fate in the gut
Selected by Marina SchernthannerCategories: bioinformatics, immunology
Updated 5 March 2025 with a postLight by Marina Schernthanner
As the keen readers and T cell aficionados among you might have spotted already, this preprint was recently published in Nature. Staying true to its (slightly altered) title – “Tissue-resident memory CD8 T cell diversity is spatiotemporally imprinted” – it expands on the question of how immune cells are regulated by their neighborhood in a tissue.
During an infection, T cells come in different flavors. Antigen-specific defense programs in T cells are initiated through priming in lymphoid tissue, but final T cell states are regulated and solidified by cell:cell interactions and environmental sensing in non-lymphoid tissues. Here, the authors picked CD8 T cells in the intestine as a model to illustrate transcriptional (and functional) “evolution” over space and time.
Riding the wave of recent spatial transcriptomic (ST) technological breakthroughs, the authors have added unbiased Visium HD ST data on mouse small intestine to their published work to complement their findings from Xenium ST profiling (~500 genes) of the murine small intestine at acute and chronic stages following viral infection. The biological insights from all of those, performed on mouse and human samples, converge neatly: A T cell’s transcriptional state strongly correlated with its spatial position along the crypt-villus and epithelial axes in the intestine.
In the preprint, the authors focused on TGFb-signaling as an example of a pathway that occurred in a gradient along the crypt-villus axis – just like several other T cell recruitment, survival and differentiation signals. In the published version, they added and highlighted the chemokines CXCL9 and CXCL10, how they directly act on incoming T cells via CXCR3 and thus promote the development of short-lived effector T cells in the lower villus. In tandem, TGFb and CXCR3-mediated signaling elegantly demonstrated the need for constitutive (TGFb) and inducible (CXCL9/10) programs that guide T cells on their differentiation trajectory within the tissue via spatially patterned signaling gradients.
In conclusion, this paper is a prime example of how to leverage state-of-the-art ST methodology to gain new insights into how our body mounts an effective immune response. By standardizing and visualizing their results in immune allocation plots, the authors showed how antigen-specific CD8 T cells take up residence in the gut in a simple, yet effective manner. Their ST data offer a rich glimpse into the transcriptional identity, spatial location and interaction partners of not only T cells, but a vast number of tissue-resident and motile cells, while also elucidating how signaling pathways can span several cell types and operate on a tissue-wide scale. Or simply put – by zooming in and out of their data, the authors of this paper are zooming in and out of a mounting immune response across space and time.
Background of the preprint
To protect us from recurring or future infections, the immune system uses a handy trick – it retains a couple of sentinels in our tissues, so-called memory T cells, that remember previous attacks and act as a first line of defense upon exposure to the same or similar harmful agents.
CD8 T cells are cytotoxic T cells, which can recognize and kill infected cells. Memory CD8 T cells, which will remember the pathogen, are generated late during an initial infection and then reside in (and surveil) non-lymphoid tissues, such as the epithelial surfaces lining the skin, lungs or gut. Tissue-resident memory CD8 T cells (TRMs) exhibit phenotypic and functional differences to their naïve and short-lived effector-like CD8 T cell counterparts. It remains poorly understood which signals and cellular environments initiate a long-lived memory fate in T cells.
Key findings of the preprint
In this study, the authors leverage state-of-the-art spatial transcriptomic profiling approaches to map the intratissue location and transcriptional identity of CD8 TRMs in the murine and human small intestine. To do so, they used a model of systemic Lymphocytic Choriomeningitis Virus (LCMV) infection in mice, which had received LCMV-specific P14 T cell receptor transgenic CD8 cells via adoptive transfer. Timepoints of analysis included several days as well as months after infection to capture CD8 T cells at effector (early) and memory (late) stages.
Spatial profiling reveals transcriptional regionalization
The distribution of CD8 T cells in the mouse small intestine upon LCMV infection was captured via Xenium spatial transcriptomics (ST), which allowed the authors to profile a set of ~500 genes via in situ probe hybridization and multiplex imaging at single cell resolution. A cell’s location within the tissue was recorded and defined along 3 axes – its distance to the nearest epithelial cell, its position along the crypt-villus axis as well as its position along the entire small intestinal tract. T cells were found to differ significantly depending on where they localized along the epithelial and crypt-villus axes. To visualize this, the authors developed so-called Immune Allocation Plots (IMAPs), which neatly showcased the dynamic migration of LCMV-specific CD8 T cells over time as well as illustrated the transcriptional heterogeneity of differentially located cells (Figure 1).
During the early effector phase following LCMV infection, P14 LCMV-specific CD8 T cells were found in the muscularis layer, from where they gradually entered the intestinal lamina propria and epithelial layers over time (Figure 1). At 90 days post-infection (the memory stage), the authors spotted two spatially segregated T cell populations – T cells rich in polyfunctional TRM genes at the villus base and T cells highly expressing effector genes at the villus tip and predominantly interspersed between epithelial cells.
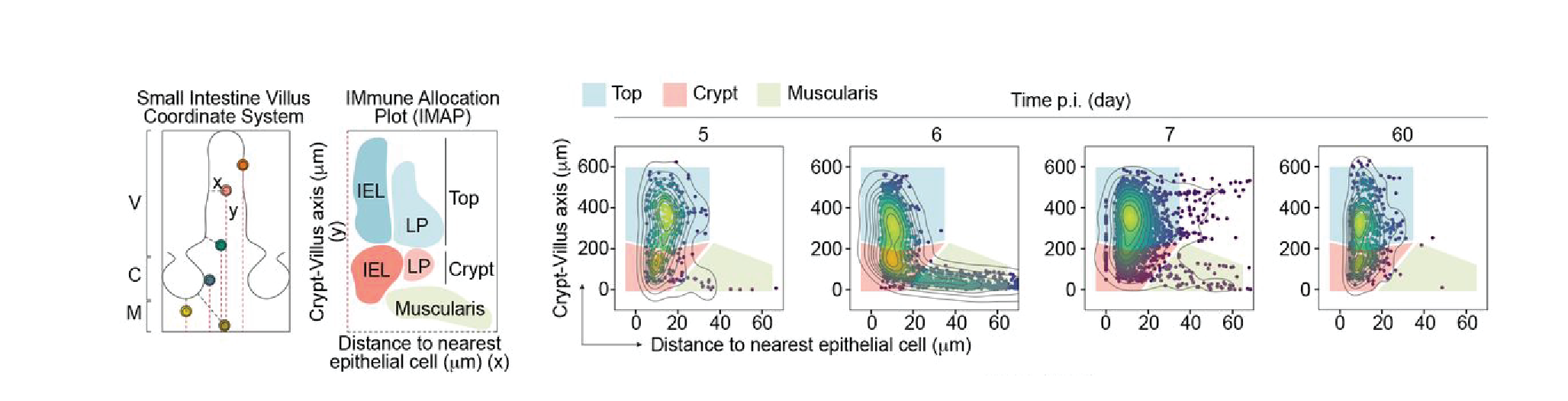
Figure 1. Mapping the transcriptional CD8 TRM fate across space and time in the murine small intestine. Immune Allocation Plots (IMAPs) illustrate the change in spatial positioning of transferred P14 T cells along the small intestinal crypt-villus axis following LCMV infection. The defined spatial domains and axes are illustrated on the left, x demarcates the distance to the nearest epithelial cells, while y denotes the distance to the base of the muscularis layer. The actual positions of T cells are mapped out across different timepoints (days post-infection (p.i.)) as detected via Xenium spatial transcriptomics on the right. Abbreviations are for villus (V), crypt (C) and muscularis layer (M).
Different interactomes and cytokine gradients along the crypt-villus axis may drive CD8 TRM heterogeneity
The differential position of the two observed TRM cell subsets already suggested a very different cellular environment for each subset. To dissect their respective interaction partners, the authors performed nearest neighbor analysis across immune, epithelial, muscularis and crypt spatial domains. Unsurprisingly, effector-like TRMs at the villus tip mostly engaged with differentiated epithelial cells (enterocytes). Polyfunctional TRMs at the villus base, in turn, appeared to crosstalk with a mix of immature epithelial cells, fibroblasts, CD4 T and B cells.
Additionally, to obtain a more comprehensive view of signaling gradients affecting immune cell dynamics, the authors performed 10X Visium spatial transcriptomics of mouse small intestines at day 30 after LCMV infection. The limited resolution of the resulting data was overcome via label transfer from Xenium spatial transcriptomics data used for deconvolution. Intriguingly, several cyto- and chemokines affecting the function and recruitment of T cells showed spatially regulated trends in expression. CXCL16 for instance peaked at the villus tip, whilst TGFb signaling was highest within the crypt.
TGFb signaling is enriched in the crypt and initiates CD8 TRM differentiation
TGFb signaling is important for the differentiation and maintenance of tissue-resident memory CD8 T cells. While the expression of TGFb receptors was relatively conserved and homogeneous in CD8 TRMs along the crypt-villus axis, TGFb ligand isoforms, mostly 2 and 3, peaked in the crypt, where they were largely expressed by fibroblasts. To test the role of TGFb signaling in instructing TRM fates, TGFbR-knockout LCMV-specific CD8 T cells were transferred into mice that were subsequently infected with LCMV. Spatial dynamics of LCMV-specific T cells were tracked via MERSCOPE at 8 days post-infection. Interestingly, TGFbR-knockout T cells were shifted towards the muscularis layer and crypt and exhibited a reduced TRM transcriptional core signature compared to wild-type T cells. Moreover, the expression of CD103, which is typically up in effector cells at the villus tip, was lower in TGFbR-knockout cells.
Differential gene expression analysis between TGFbR-knockout and wild-type T cells revealed 8 genes that were downregulated in knockout cells. Mapping this gene signature back onto the wild-type spatial transcriptomics data revealed its preferential expression in villus tip T cells. In conclusion, ablation of TGFb signaling in LCMV-specific CD8 T cells appears to impair their differentiation into tissue-resident memory CD8 T cells and abrogate their spatial trajectory along the crypt-villus axis.
Conclusions
By leveraging spatial transcriptomics approaches, the authors of this preprint profiled the murine and human small intestine in spatial detail. In doing so, they uncovered how and where virus-specific CD8 T cells following infection with LCMV adopt effector- versus memory-like fates over time. At the memory stage, they characterized two transcriptionally distinct states of CD8 T cells, which resided in different tissue niches. Importantly, those two subsets arose in wild-type host T cells and human tissue as well, strongly suggesting that spatial imprinting is a physiological process. T cells lacking the receptor for TGFb-signaling exhibited impaired differentiation towards a TRM state during early timepoints after LCMV infection. This highlights TGFb signaling, which was concentrated in crypt regions, as an important candidate signaling pathway in TRM fate acquisition (Figure 2).
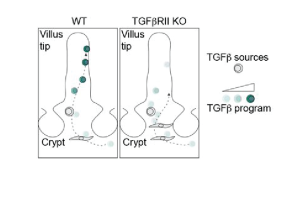
Figure 2. Model of fibroblast-derived TGFb-mediated effector TRM maturation along the crypt-villus axis in the intestine.
What I like about this preprint
As the reader can infer from the extensive supplement, this manuscript serves as a great resource to probe spatially regulated transcriptional signatures in multiple cell types. It highlights the power and information that state-of-the-art spatial transcriptomic approaches offer to finally shed light on how the environment affects a cell’s identity and function. This becomes particularly interesting when looking at motile cell types, such as immune cells, which may transit through several different microenvironments as they adopt their fate.
Here, Reina-Campos, Monell and colleagues indeed propose a gradual TRM differentiation trajectory as the cells travel from crypt to villus upon entry into the tissue that (to some extent) depends on TGFb signaling. It sparks the notion of a multi-step differentiation process – a general (“inborn”) gradual fate acquisition as cells pass through several signaling environments from crypt-to-villus, complemented by final fate determination depending on which cells they end up interacting with. Whether this concept extends to other cell types, especially motile ones, remains to be explored but opens doors to exciting new questions.
Future directions and questions for the authors
Given the use of an infection model and spatial transcriptomics profiling across several timepoints, the findings of this paper amount to an interesting spatiotemporal atlas. This allows glimpses into the process of T cell (here CD8 TRM) differentiation in never-before-seen detail and unsurprisingly, I am left intrigued and with a lot of questions:
- The interactomes of effector-like TRMs at the villus tip versus polyfunctional TRMs at the villus base are quite different, also in terms of cell type heterogeneity. Villus base TRMs seem to interact with many more cell types than villus tip TRMs. Could (presumably very different) signals from multicellular sources favor a polyfunctional TRM fate at the villus base over a more specialized effector-like fate as seen in villus tip TRMs, which predominantly seem to interact with enterocytes?
- Several signals involved in immune cell recruitment (Madcam-1, chemokines etc.) appear to peak in the crypt region. Why would passing by the crypt on their way into the tissue be the default mode for motile immune cells? Besides TGFb ligands, did the authors look into additional signals enriched in the crypt that might be relevant for immune cell maturation? Or alternatively, could signals like WNTs, which are known to be high in the crypt and relevant for epithelial stem cell maintenance, inevitably affect incoming immune cells as well and if so, how?
- Molecules like IL-10 appear to be enriched in the crypt region and in general this spatial domain appears to be more immunosuppressive and enriched in progenitor-like cells (across epithelial and immune cell types) and polyfunctional memory-like cells. A functional and spatial segregation into effector and progenitor/memory cells at the villus tip versus bottom/crypt intuitively makes sense – antigen and pathogen exposure predominantly occurs in villi, whilst barrier-renewing stem cells sit in the crypt and need to be shielded from immune attacks. But do the authors think that this segregation might actually be driven by and mimic epithelial differentiation processes along the crypt-villus axis?
- The spatial shift of TGFbR-knockout T cells towards the crypt is interesting. Given the authors’ focus on TRMs, it would have been interesting to also include a later (memory) timepoint following adoptive transfer of TGFbR-knockout T cells into LCMV-infected mice.
- Could the shift of knockout T cells towards the crypt just be a reflection of a delayed TRM program? Would TGFbR-knockout T cells eventually be able to migrate towards the villus tip at a later timepoint?
- Is virus clearance affected/delayed in mice that were injected with TGFbR-knockout cells? And are they more susceptible to a recurring infection?
- Given the role of TGFb signaling in TRM maintenance – did the authors observe differences in the lifespan of TGFbR-knockout versus control T cells?
- I wonder whether the authors have given any thought to whether these processes are conserved during early postnatal life, when mice (or humans) start to become exposed to a lot of different antigens and have to mount effective immune responses for the first time?
- Did the authors look into transcriptional heterogeneity of any other immune cell types and were they able to delineate spatially segregated populations in a similar fashion?
doi: https://doi.org/10.1242/prelights.38184
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the bioinformatics category:
Decoding the Molecular Language of Proteins with Evolla
Jawdat Sandakly
Tidyplots empowers life scientists with easy code-based data visualization
Felipe Del Valle Batalla
IMMClock reveals immune aging and T cell function at single-cell resolution
Jessica Chevallier
Also in the immunology category:
Discovery of a Potent and Selective Inhibitor of Human NLRP3 with a Novel Binding Modality and Mechanism of Action
Roberto Amadio
Target cell tension regulates macrophage trogocytosis
Felipe Del Valle Batalla
Leukocytes use endothelial membrane tunnels to extravasate the vasculature
Felipe Del Valle Batalla
preLists in the bioinformatics category:
‘In preprints’ from Development 2022-2023
A list of the preprints featured in Development's 'In preprints' articles between 2022-2023
| List by | Alex Eve, Katherine Brown |
9th International Symposium on the Biology of Vertebrate Sex Determination
This preList contains preprints discussed during the 9th International Symposium on the Biology of Vertebrate Sex Determination. This conference was held in Kona, Hawaii from April 17th to 21st 2023.
| List by | Martin Estermann |
Alumni picks – preLights 5th Birthday
This preList contains preprints that were picked and highlighted by preLights Alumni - an initiative that was set up to mark preLights 5th birthday. More entries will follow throughout February and March 2023.
| List by | Sergio Menchero et al. |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |
Also in the immunology category:
Community-driven preList – Immunology
In this community-driven preList, a group of preLighters, with expertise in different areas of immunology have worked together to create this preprint reading list.
| List by | Felipe Del Valle Batalla et al. |
Journal of Cell Science meeting ‘Imaging Cell Dynamics’
This preList highlights the preprints discussed at the JCS meeting 'Imaging Cell Dynamics'. The meeting was held from 14 - 17 May 2023 in Lisbon, Portugal and was organised by Erika Holzbaur, Jennifer Lippincott-Schwartz, Rob Parton and Michael Way.
| List by | Helen Zenner |
Fibroblasts
The advances in fibroblast biology preList explores the recent discoveries and preprints of the fibroblast world. Get ready to immerse yourself with this list created for fibroblasts aficionados and lovers, and beyond. Here, my goal is to include preprints of fibroblast biology, heterogeneity, fate, extracellular matrix, behavior, topography, single-cell atlases, spatial transcriptomics, and their matrix!
| List by | Osvaldo Contreras |
Single Cell Biology 2020
A list of preprints mentioned at the Wellcome Genome Campus Single Cell Biology 2020 meeting.
| List by | Alex Eve |
Autophagy
Preprints on autophagy and lysosomal degradation and its role in neurodegeneration and disease. Includes molecular mechanisms, upstream signalling and regulation as well as studies on pharmaceutical interventions to upregulate the process.
| List by | Sandra Malmgren Hill |
Zebrafish immunology
A compilation of cutting-edge research that uses the zebrafish as a model system to elucidate novel immunological mechanisms in health and disease.
| List by | Shikha Nayar |











 (No Ratings Yet)
(No Ratings Yet)