Diverse logics and grammar encode notochord enhancers
Posted on: 21 August 2022
Preprint posted on 27 July 2022
Article now published in Cell Reports at http://dx.doi.org/10.1016/j.celrep.2023.112052
If transcription factors provide the vocabulary for development, what are the grammatical rules by which they’re interpreted? @EKFarley and colleagues use the sea squirt notochord to study the logic and grammar of enhancers driving the developmental
Selected by Andrew MontequinBackground
One of the mysteries of developmental biology is how the entire diversity of cell types found in an adult organism is specified by a relatively small set of signaling molecules and transcription factors. While single transcription factors can play multiple distinct roles in development, combinations of transcription factors can encode increasingly complex signals guiding cell fate decisions (Levine & Tjian, 2003). During development, cells act as semi-autonomous machines that interpret combinatorial input signals and execute a specific program to adopt the correct fate. When this “program” runs without error, the result is a complex body plan that has the right cell types in the right place.
A large body of work in developmental biology has focused on understanding when and where transcription factors are expressed (Ilsley et al., 2020; Winkley et al., 2021), a task akin to understanding the inputs to the developmental program. Less attention has been paid to how these input signals are interpreted. Enhancers, which are genomic elements rich in transcription factor binding sites, serve to organize these combinatorial signals to drive tissue- and stage-specific gene expression. The grammar of these enhancers – including the number, spacing, orientation, and affinity of transcription factor binding sites – is thought to play a significant role in regulating the activity of the enhancers.
Previous research by Dr. Emma Farley, now the corresponding author on this preprint, uncovered some of the grammatical rules governing notochord specification (Farley et al., 2016). In this preprint, Song and colleagues use the sea squirt Ciona to dive deeper into how combinations of the transcription factors Zic, ETS, FoxA and Bra are organized at notochord enhancers. In doing so, they uncover conservation in enhancer grammar across chordates, suggesting that universal grammatical constraints on enhancers may exist in the animal kingdom.
Main Findings
A subset of Zic and ETS genomic elements are enhancers driving notochord expression
Building on previous work that identified low-affinity ETS binding sites as necessary for notochord development (Farley et al., 2016), the authors mined the Ciona genome for elements containing multiple ETS sites in proximity to Zic binding sites. Out of 1092 identified ZEE elements, or genomic regions containing at least one Zic site and two ETS sites, 90 were selected for further study. Using an assay to identify potential enhancers, fewer than half of these elements appeared sufficient for driving gene expression anywhere in the embryo and only nine were sufficient for driving expression in the notochord.
Proper Zic and ETS grammar is necessary but not sufficient for notochord activity
One of the putative notochord enhancers was found to be located close to laminin alpha, a gene that is critical for notochord development. Point mutations to the Zic site and the highest-affinity ETS site, as well as reversal of that same ETS site, all eliminated the ability of this region to drive gene expression. Further studies into a previously characterized notochord enhancer that drives brachyury expression similarly found that point mutations to Zic, ETS, Bra or FoxA sites all disrupted gene expression. Randomizing the base pairs outside of the Zic and ETS sites for this enhancer also disrupted gene expression, indicating that Zic and ETS sites are necessary but not sufficient to drive notochord expression (Fig. 1).
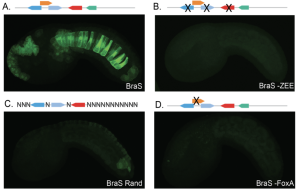
Figure 1: (A) A reporter for the BraS enhancer drives GFP expression in the notochord. (B) Point mutations in the Zic and ETS sites within the BraS enhancer render the enhancer non-functional. (C) Randomizing the base pairs outside of the BraS site render the enhancer non-functional, indicating that there are necessary elements in addition to Zic and ETS sites within the enhancer. (D) A point mutation to the FoxA site renders the enhancer non-functional. Figure adapted from preprint Figure 5 A-D.
Enhancer logic may be conserved across chordates
The authors identified Zic and ETS clusters within intronic regions of the mouse and human laminin alpha-1 gene, providing evidence that components of the laminin alpha enhancer discovered in Ciona may be conserved across chordate species. Several striking similarities were observed across all three species, including a 12 base pair distance between adjacent Zic and ETS sites as well as the appearance of non-consensus ETS sites within these regions. Comparisons between brachyury enhancers across Ciona, mouse and human identified clusters of Zic, ETS, FoxA and Bra sites within all species, indicating that there may be a shared regulatory logic within notochord enhancers.
Why I chose this preprint
This preprint gives an important perspective on the complexity of signal integration in developing organisms. While so much of developmental biology is rightfully focused on understanding when and where sets of transcription factors are expressed, this work serves as a reminder that a narrow focus on expression patterns provides an incomplete understanding of development. The authors of this preprint expertly build on previous expression studies to begin to understand how co-expression of transcription factors may be interpreted to influence cell fate decisions.
Beyond the results, I felt that this preprint was very well-written. The authors provide a background on enhancer biology that clearly and succinctly explains the theory of enhancer “grammar” and the importance of understanding this grammar. I was easily able to grasp the experimental set up, data, and relevance of the work despite having a limited background in this sub-field. Ciona embryos provide an experimentally tractable system for these experiments, and the authors did a fantastic job of connecting their results to deeper principles that may be shared across the animal kingdom. I never felt like I was reading a paper where the results would only be applicable to a single organism.
Questions for the Authors
- When you grouped the 90 ZEE elements you studied based on their proximity to identified Bra and FoxA sites, each group varied in size. For example, 39 elements contained Zic/ETS/Bra sites, but only 5 contained Zic/ETS/FoxA sites. If you performed the same analysis on all 1092 ZEE elements, would you expect to see a similar distribution of group sizes?
- I’m interested in how enhancer logic may tune transcriptional outputs in the presence of noisy signal inputs, such as single-cell fluctuations in transcription factor concentrations. In addition to the Lama mutations in the preprint that completely ablated GFP expression, are you aware of changes to the enhancer grammar that lead to more heterogeneous expression?
References
Farley, E. K., Olson, K. M., Zhang, W., Rokhsar, D. S., & Levine, M. S. (2016). Syntax compensates for poor binding sites to encode tissue specificity of developmental enhancers. Proceedings of the National Academy of Sciences, 113(23), 6508–6513. https://doi.org/10.1073/pnas.1605085113
Ilsley, G. R., Suyama, R., Noda, T., Satoh, N., & Luscombe, N. M. (2020). Finding cell-specific expression patterns in the early Ciona embryo with single-cell RNA-seq. Scientific Reports, 10(1), 4961. https://doi.org/10.1038/s41598-020-61591-1
Levine, M., & Tjian, R. (2003). Transcription regulation and animal diversity. Nature, 424(6945), 147–151. https://doi.org/10.1038/nature01763
Winkley, K. M., Reeves, W. M., & Veeman, M. T. (2021). Single-cell analysis of cell fate bifurcation in the chordate Ciona. BMC Biology, 19(1), 180. https://doi.org/10.1186/s12915-021-01122-0
doi: https://doi.org/10.1242/prelights.32555
Read preprint










 (No Ratings Yet)
(No Ratings Yet)