Gain of gene regulatory network interconnectivity at the origin of vertebrates
Posted on: 3 June 2020
Preprint posted on 27 April 2020
Article now published in Proceedings of the National Academy of Sciences at http://dx.doi.org/10.1073/pnas.2114802119
In evolution, more is more: Higher interconnectivity among developmental pathways and enrichment of highly connected genes in vertebrate novelty tissues – clues on the origins of vertebrates.
Selected by Glò Casas GimenoIntroduction
Cephalochordates and vertebrates diverged from their common chordate ancestor over 520 million years ago [1]. From then, vertebrate evolution has been characterized by the appearance of novel cell types and tissues, like adaptative immunity, and the neural crest and its most famous derivative, the endoskeleton [2]. How did the already complex developmental mechanisms evolve to give rise to these new features is a fundamental concern of evo-devo. At the genomic level, two whole genome-duplication events at the apex of the vertebrate lineage laid favorable grounds for evolution. Rather than acquiring completely new functions, most duplicated developmental genes maintained their developmental involvement [3]. At the same time, gene regulatory landscapes for these genes became more intricate.
In order to explore the precise role of interconnectivity of gene regulatory networks in the invertebrate-to-vertebrate transition, Gil-Gálvez and colleagues use a simple (in the best sense of the term) experimental set-up. Taking the cephalochordate amphioxus* and zebrafish as invertebrate and vertebrate models, respectively, they use omics to compare how four major developmental signaling pathways (Wnt, Nodal, FGF and RA) interact with each other. The figure below, from Figure 4. in the pre-print, speaks for itself: these pathways are more strongly interconnected in the vertebrate zebrafish than the invertebrate amphioxus.
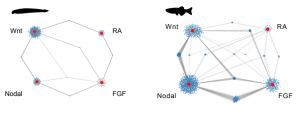
*For a beautiful overview of amphioxus development, check the pre-print by Carvalho et al. (https://doi.org/10.1101/2020.05.26.112193) [4]
Research summary
Briefly, amphioxus and zebrafish embryos were treated with chemicals that perturb the selected signaling pathways (activators of Wnt and RA, and inhibitors of Nodal and FGF). Embryos were collected at late gastrula stage for RNA seq and ATAC-seq analysis.
For FGF and Nodal, there was a big overlap in differentially detected transcripts upon treatment in the two species. Interestingly, this overlap was notably smaller in the case of RA, and especially in the case of Wnt treatment.
In comparison, a relatively higher number of genes was altered in zebrafish than in amphioxus in response to all individual treatments. This was true both at the transcript level and by ATAC-seq analysis of open chromatin regions.
Clustering analysis of both ATAC-seq data and RNA-seq data also show that most of the identified gene clusters were responsive to more than one treatment, especially in vertebrates. When comparing how many genes are regulated by one, two, three or four of the pathways, they found that zebrafish genes were much more commonly affected by two or more manipulations than amphioxus genes. Altogether, these analyses coincide with a higher degree of interconnectivity in zebrafish than in amphioxus.
Finally, the last figure of the main text illustrates an intriguing concept: novel vertebrate tissues, like the neural crest, express a higher proportion of “highly connected genes” (i.e. responsive to ≥ 3 of the treatments), whereas tissues of ancient evolutionary origin, like the intestine, show a similar expression of “highly connected” and “lowly connected” genes (i.e. responsive to 1 treatment). This is suggestive of a direct link between gain of connectivity and emergence of tissue innovations in vertebrate evolution.
Questions for the authors
- I am curious about the difference between FGF and Nodal compared to Wnt and RA in terms of the overlap of differentially expressed genes in amphioxus and zebrafish. Do you think it is reflective of the level of conservation of these pathways, or do you favor another explanation?
- In your first ATAC-seq analysis, you look at genes that are more accessible upon treatment with Wnt or RA agonist and less accessible upon treatment with either FGF or Nodal antagonist. Why did you choose to not include all genes that show differential accessibility, as you do in later analyses?
- This is a very naïve conceptual question: is connectivity in gene regulatory network maintained across tissues? If no, do your results support that there is a higher connectivity in novelty tissues than in more ancestral ones?
References
[1] Holland LZ et al. The Amphioxus Genome Illuminates Vertebrate Origins and Cephalochordate Biology. Genome Res. 2008 Jul;18(7):1100-11.
[2] Shimeld SM, Holland PW. Proc Natl Acad Sci U S A. 2000 Apr 25;97(9):4449-52.
[3] Marlétaz F et al. Amphioxus functional genomics and the origins of vertebrate gene regulation.Version 2. Nature. 2018 Dec;564(7734):64-70.
[4] Carvalho et al. An updated staging system for cephalochordate development: one table suits them all. bioRxiv 2020 May.
doi: https://doi.org/10.1242/prelights.21513
Read preprint










 (No Ratings Yet)
(No Ratings Yet)