In vivo identification of GTPase interactors by mitochondrial relocalization and proximity biotinylation
Posted on: 1 March 2019 , updated on: 5 March 2019
Preprint posted on 17 February 2019
Article now published in eLife at http://dx.doi.org/10.7554/elife.45916
Artificial localization of small GTP binding proteins to mitochondria enables proteomic identification of new interactions for Rabs and other superfamily members.
Selected by David StephensBackground
Small GTPases control diverse cellular processes including membrane trafficking and cytoskeletal dynamics. They are activated by guanine nucleotide exchange factors, bind to a series of effector molecules, and are inactivated following GTP hydrolysis stimulated by GTPase activating proteins. Work to identify small GTP binding protein effectors has exploited many protein-protein interaction technologies over the years, notably yeast two-hybrid methods and affinity purification. Here, the Munro lab has developed and applied mitochondrial relocation coupled with spatially-resolved proteomics to identify proteins that interact with Rab GTPases. The authors also show broader application of the method using key cytoskeletal regulators RhoA, Rac1, and Cdc42.
Key findings
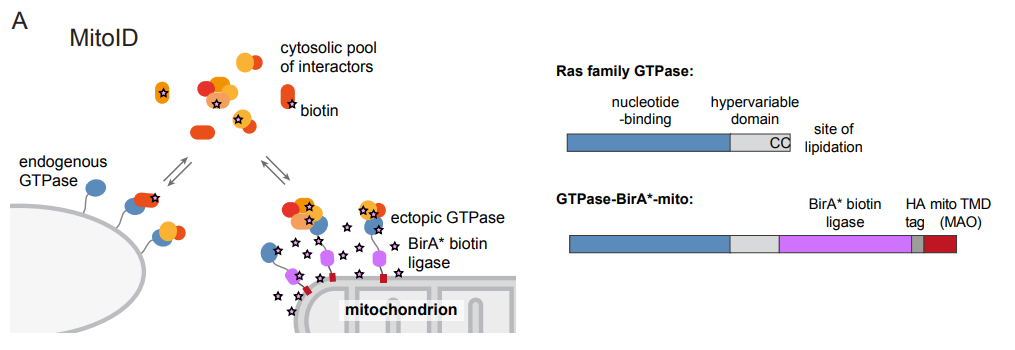
In the current study, Alison Gillingham, Jessie Bertram and colleagues applied biotin-based proximity labelling (BioID) approach using a promiscuous BirA* biotin ligase fused to the GTPase of interest. This fusion construct was targeted to the mitochondria using the transmembrane domain of monoamine oxidase to provide an artificial but tightly restricted mitochondrial localization of this fusion construct. Following biotinylation, proteins are isolated by streptavidin affinity capture and analysed by mass spectrometry. This MitoID approach was validated using GTP- and GDP-restricted forms of Rab5A, which has a well-defined set of interacting partners. Nearly all expected interactions were detected along with several new candidates. Application of the method to 10 other Rabs as well as other small GTPases (Cdc42, RhoA, Rac1, Rheb, RalA, and N-Ras) identified many new candidate interactors as well as most existing ones. Some key and unexpected findings from the work include the identification of many centrosomal and cilia proteins interacting with Rab2A, many new Rab-motor protein interactions, and as one might expect, many golgins. Interesting “nuggets” within the data set include identification of MYO18A and kinesin-2 KIF3A/B with Rab10, a role for Rab9 in the recruitment of the dynein motor regulator NDE1 to endosomes, and links between RhoA and the dynein adaptor BICD.
Perspective
Proximity-based labelling of proteins coupled with proteomic identification is now a key methodology for mechanistic cell biology research (Kim and Roux, 2016). It has some advantages over previous approaches and can readily be undertaken in living cells. A key issue with any of these proximity labelling technologies is the identification of “bystanders” rather than bona fide interactors. By retargeting small GTPases to the mitochondrial outer membrane, this problem was reduced. A consistent set of background interactions was detected, and a series of specific regulators and effectors defined. A particular benefit is that this enables the direct comparative analysis of GTP- and GDP-bound small G-proteins, the latter not normally being membrane-associated. The method developed doesn’t use particularly large numbers of cells, or require the generation of stable lines, enhancing the likelihood of its implementation more widely. This work will be of interest to those working on the many proteins that have been defined through this interaction analysis, as well as to those looking to develop this technique for their own ends. Potential pitfalls broadly overlap with those relevant to any other similar approach. Further questions would include whether there is any preferential outcome with regard to the interactions detected. For example, one could envisage that the method artificially clusters the GTPases such that lower affinity interactions might be under-represented. The location of the fusion domains, despite careful placement of linkers, could compromise some key interactions. Other multi-partite interactions might be missed where membrane context is important (lipid composition, curvature, colocation of other essential components). It is important to bear in mind that any similar approach suffers from similar limitations and that does not preclude effective implementation of the approach. Careful experimental design, extensive validation, and considered interpretation will always be necessary.
Why I like this preprint
This work provides an elegant example of how combining existing methods (here, artificial mitochondrial targeting and proximity labelling) can add significant value. I also like the fact that this new discovery step is applied and validated, not only for the Rab GTPases, but also for other important and widely studied enzymes. I also very much like the way that this manuscript defines extensive new biological insight that goes far beyond a simplistic description of the method. This gives real confidence in terms of adopting this oneself. One can easily envisage broad and diverse applications of the method.
Open questions
Could the method be further improved through the use of newer biotin ligase systems (Branon et al., 2018)?
How readily could this approach be applied to other small GTPases such as the Arf family, to other nucleotide-switched systems such as motor proteins?
Could this technique be used in more complex in vivo scenarios to probe developmental or tissue-specific difference in interactomes?
What are the biological functions of these newly identified interactions?
Does Rab2A have a role at the centrosome or in ciliogenesis?
References
Branon, T. C., Bosch, J. A., Sanchez, A. D., Udeshi, N. D., Svinkina, T., Carr, S. A., Feldman, J. L., Perrimon, N. and Ting, A. Y. (2018). Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol 36, 880-887.
Kim, D. I. and Roux, K. J. (2016). Filling the Void: Proximity-Based Labeling of Proteins in Living Cells. Trends Cell Biol 26, 804-817.
doi: https://doi.org/10.1242/prelights.9097
Read preprint










 (No Ratings Yet)
(No Ratings Yet)