Interactive design of GPU-accelerated Image Data Flow Graphs and cross-platform deployment using multi-lingual code generation
Posted on: 20 December 2020
Preprint posted on 20 November 2020
Background
Modern research in life sciences relies heavily on fluorescence microscopy, and subsequent quantitative image analysis. The availability of image analysis algorithms to a broader audience boosts the need for accessible tools for building graphics processing units (GPU)-accelerated image analysis workflows. The rise of GPUs in the context of image processing enables batch processing large amounts of image data at unprecedented speed. Usually, designing data analysis procedures using GPUs requires expertise in programming, and knowledge of GPU-specific programming languages. Haase and colleagues (1) present here an expert system based on the GPU-accelerated image processing library CLIJ. They hereby demonstrate the construction of complete image analysis pipelines by assembling workflows from operations provided by the CLIJ framework. The user interface CLIJ-assistant allows interactive design of image data flow graphs (IDFGs) in ImageJ or Fiji, while guiding the user by keeping track of which operations formed an image, and suggesting subsequent operations. Operations, their parameters and connections in the IDFG are stored at any point in time enabling the CLIJ-assistant to offer an undo-function for virtually unlimited rewinding parameter changes. The CLIJ-assistant can generate code from IDFGs in multiple programming languages for later use in multiple image analysis platforms. The CLIJ-assistant is open source, and available online at https://clij.github.io/assistant/
Key findings and developments
Key developments
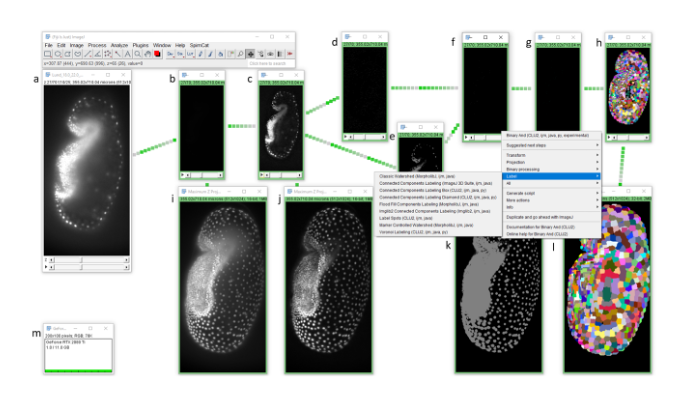
Design. CLIJ-assistant was implemented under the ImageJ user-interface, to improve accessibility by users. In the background, CLIJ-assistant manages an IDFG. The image data are propagated in the graph in one direction only, without loops being constructed. The approach used is similar to the user interfaces of Icy and Knime, with the advantage that intermediate results of the whole graph are always images which can be updated instantly in ImageJ’s user interface, while the user changes parameters. To optimize performance, the image stacks are kept in the GPU’s memory while minimizing pushing and pulling image data to/from GPU memory. To visualize the state of a graph, IDFG windows appear differently than standard ImageJ windows by their frame colour, visualizing the graph’s execution state: a red frame indicates that the shown image is invalid and will be computed when intermediate results higher in the graph hierarchy are available. A yellow frame shows a currently ongoing computation. A green frame indicates a computation is finished. Upon moving a window, all downstream windows also move, thus giving the user an impression of connections between graph nodes. This way of user interaction with the imaging data allows users to learn image processing and analysis, and the relationship between operations more efficiently, as it keeps technical implementation details out of sight.
For every processed image, CLIJ-assistant can backtrack, so that it is not necessary to store intermediate results of various parameters configurations in the computer memory. The possibility of rewinding to former parameter settings brings a virtually unlimited undo functionality to Fiji. Parameter changes can be documented within and between projects.
Available operations and extensibility. Following installation of the CLIJ-assistant, about 249 image processing operations become available. The CLIJ-assistant uses ImageJ2 plugin mechanisms to automatically discover additional CLIJ-compatible plugins, therefore, custom third-party operations can be introduced as graph nodes.
Expert system. Image analysis workflows can be thought of as an assembly of operations. In recent years, in order to guide users, a search-bar and an auto-completion of commands in the script editor were introduced to facilitate accessibility and application of the steps in the image analysis workflow. However, neither the search-bar nor the auto-completion tools suggest what to do next. To overcome this possible limitation, the authors implemented an expert system in the CLIJ-assistant. Expert systems are a form of artificial intelligence developed to for instance, help users in the decision making process. In this case, the expert system guides users in choosing the right operations step by step, by making context-dependent suggestions based on previously executed operations. Nevertheless, all operations are available under all circumstances from Fiji’s search bar, giving the user full access to all available CLIJ-assistant compatible operations.
Semi-automated parameter optimization. The authors introduce a simple annotation tool based on ImageJ’s ROI manager and an automatic parameter optimization tool. It is recommended to start the optimization with a good manual initial guess.
Code generation for automation, documentation and knowledge exchange. The authors implemented code generation capabilities in the CLIJ-assistant using an IDFG as a starting point. After an IDFG has been set up, configured, and optimized, the graph can be exported in various programming languages. Moreover, it is possible to compare scripts in multiple languages. The exported scripts allow the user to go beyond ImageJ and Fiji because they also offer programming languages applicable in other platforms. Supported languages are ImageJ Macro, Icy, Javascript, Matlab, Fiji Groovy, JavaScript, and Fiji Jython. Meanwhile, support for Python and C++ are under development, allowing prototype testing. In order to foster reproducibility of image analysis procedures by clear documentation, human-readable protocols of the IDFG can be exported. These protocols can be used to communicate the applied image processing workflow with scientists using other platforms, or people without coding experience.
Key applications
The authors demonstrate the capabilities of CLIJ-assistant in four different contexts, namely gut neuroscience, developmental biology, and cancer research.
Gut neuroscience context: Interactions between the enteric nervous system and the resident immune cells are important for the normal functioning of the gut. In disease, the spatial organization of these cells can be disrupted. The authors used a combination of LSFM and optical clearing to acquire images of an optically cleared mouse colon in which different structures were fluorescently labeled. The challenge of this dataset is the separation of the different layers of the gut within the image, and the ability to view cell types within each layer. The authors developed and demonstrated a workflow with multiple steps to answer the question on spatial organization of cells in the gut in health and disease.
Developing Tribolium embryos: Gastrulation is a major developmental event in an organism’s life. 2D cell shape changes and 3D volumetric shape changes are observed in tissues in an embryo as they acquire their final shapes. The authors developed and demonstrated a workflow to understand the contribution of cell behaviours to tissue morphogenesis in developing embryos. The authors also investigated Tribolium embryo development upon digital serosa removal. Serosa is an outer extra-embryonic protective layer, with different cell shapes and mechanical properties along the dorsal-ventral axis. The authors developed an IDFG to identify and selectively digitally remove the serosa.
Cell classification on 2D histological mouse brain sections. The main question in these sections was to investigate how anti-cancer treatments affect both the tumour itself but also the surrounding healthy tissue. A reporter mouse line offers the possibility to study the effects of radiation on cells, including investigating the damaged cell fraction, and how it correlated with the applied radiation dose.
What I like about this preprint
I like the fact that CLIJ-assistant offers huge versatility and applicability, and that the authors actively encourage users to take part in open calls for contributions. I think as a development this is a fantastic tool for the scientific community, including everyone using microscopy who might have been so far limited if coding knowledge was missing. CLIJ-assistant is consistent with the philosophy of democratizing science, and producing open science.
References
- Haase et al, Interactive design of GPU-accelerated image data flow graphs and cross-platform deployment using multi-lingual code generation, bioRxiv, 2020.
doi: https://doi.org/10.1242/prelights.26574
Read preprint










 (No Ratings Yet)
(No Ratings Yet)