Programmable CRISPR interference for gene silencing using Cas13a in mosquitoes
Posted on: 12 December 2019 , updated on: 11 February 2021
Article now published in Journal of Genomics at http://dx.doi.org/10.7150/jgen.43928
CRISPR-Cas13 is the new Cas9: A new and improved version of gene editing and expression manipulation
Selected by Yasmin LauBackground
The ability to manipulate gene expression has unfolded from an option into an integral asset in modern molecular biology. Today, this is achieved with a repertoire of techniques that can be performed on a number of model organisms. Cleverly, these techniques mostly exploit existing machinery within defence and adaptive immune responses in both prokaryotes and eukaryotes, such as the CRISPR (clustered regularly interspaced short palindromic repeats)-cas9 system and RNA interference.
RNA interference in eukaryotes is triggered by the introduction of virus double-stranded RNA. This defensive response ultimately entails gene silencing which involves an Argonaut nuclease-containing RISC complex (RNA-inducible silencing complex) coupled with Dicer (a ribonuclease III enzyme) (1). Together, double-stranded RNA is recognised and cleaved into small-interfering RNA (siRNA) and the target gene on the mRNA transcript is silenced (1). In a similar fashion, the CRISPR-cas9 system in prokaryotes also functions as a response to invading virus DNA and RNA. The nuclease cas9 recognises target DNA using the CRISPR RNA array (crRNA) which encodes the guide RNA. The entire system consists of Cas genes and repetitive elements interspaced by variable sequences called protospacers (2). These protospacers are also associated with a short protospacer adjacent motif (PAM), which varies with the specific CRISPR system (3). Cas9 is thus encoded, associated with the guide RNA and subsequently directed by each unit of the crRNA with a 20-nucleotide base pair sequence via Watson-Crick base pairing (2), where the target DNA is cleaved.
Recently, a new genome-editing CRISPR-Cas13 system which acts on the mRNA level has been tested to focus efficiency and reduce off-target collateral effects. In this preprint, the authors describe this method to monitor vector competence in mosquitos by probing the expression of Cas13 from an engineered construct and, subsequently, its efficiency in silencing target genes.
Key Findings
Expression of the Cas13a construct
The authors first constructed a modified plasmid containing the Cas13a gene attached with a constitutive promoter and expressed transcripts were detected using RT-PCR. This was done by injecting the plasmid into the thorax of An. gambiae and Ae. aegypti mosquitos. In An. gambiae, RT-PCR of Cas13a and rpS5 (loading control) was carried out 3 days after injection on both mosquitos with and without the injection (Figure 1).
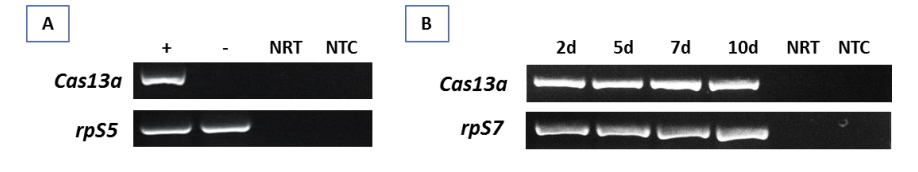
Figure 1 – RT-PCR of Cas13a and rpS5 in A) An. gambiae after 3 days and B) Ae. aegypti after 2, 5, 7 and 10 days
rpS5 was detected in both mosquitos with and without injection, while the Cas13a transcript was only present in the former. The Cas13a transcript was also detected at 2, 5, 7 and 10 days after injection in Ae. aegypti. No bands were visible for the negative controls (No RT control and no template control).
Gene silencing of Vitellogenin and COPI genes in An. gambiae and Ae. aegypti
Next, the authors tested the efficacy of gene silencing by Cas13a. The genes selected were Vitellogenin, a yolk protein precursor essential for oogenesis after blood feeding (4), and the COPI (coatomer complex I) gene, which encodes for proteins involved in blood digestion in Ae. aegypti (5). In An. gambiae, the Cas13a construct was introduced 3 days prior to blood meal-induced Vg expression. Vg proteins are found in developing oocytes during oogenesis. A control crRNA or Vg crRNA was then injected into the females 2 hours after feeding to induce Vg knockdown. The frequency of Vg transcripts was reflected in each group from the number of eggs produced, where this was reduced in those injected with the Vg crRNA (Figure 2).
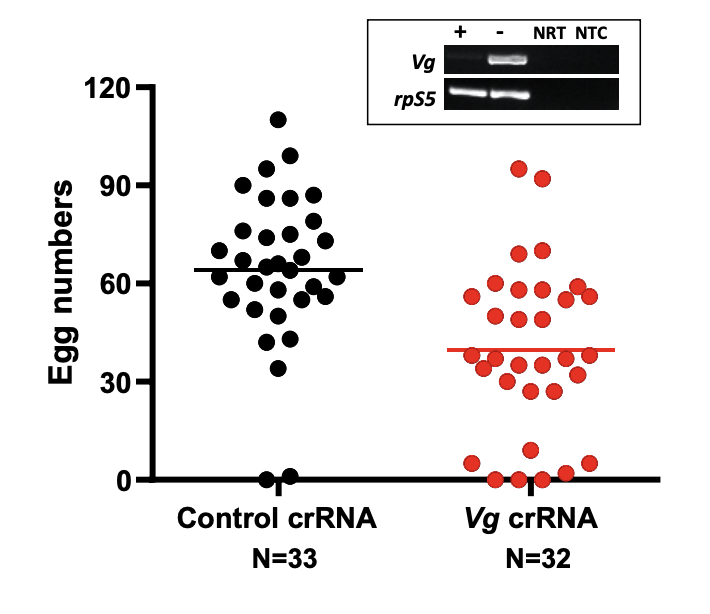
Figure 2 – Frequency of egg production in female An. gambiae mosquitos injected with control crRNA or Vg crRNA
In mosquitos, each COPI protein subunit (7 in total) is encoded for by a separate COPI gene. Knockdown of the COPI genes leads to blood meal-induced mortality (5). In Ae. aegypti, silencing of the COPI genes was tested by injecting the Cas13a construct into the mosquitos, and a blood meal was given to them after 3 days. These were subsequently divided into 2 groups, one with the a and d COPI crRNAs injections and the other injected with control crRNA. To measure the efficacy of COPI gene knockdown in each group, survival over 9 days was measured as a phenotype of blood meal-induced mortality (Figure 3).
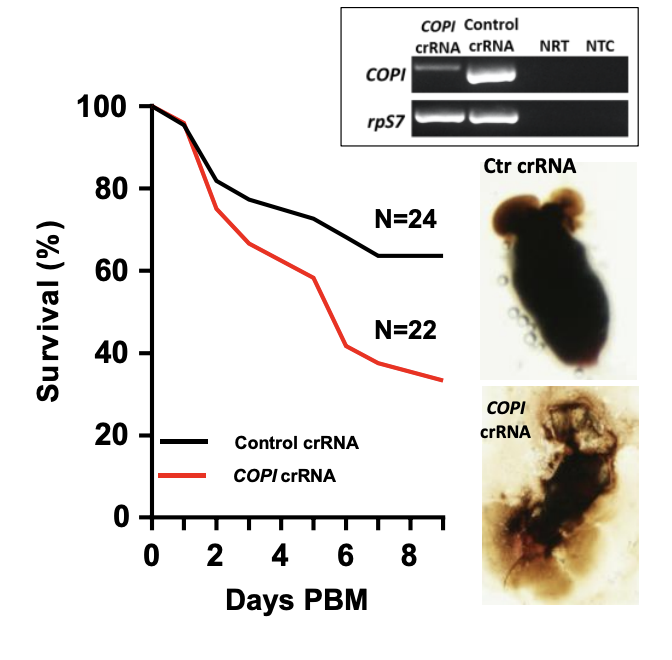
Figure 3 – Survival curves of mosquitos injected with COPI crRNA and control crRNA over 9 days (PBM = post-blood meal)
A reduction of COPI transcripts was observed in mosquitos injected with the COPI crRNA, which was reflected in their higher mortality rate compared to the control group. COPI transcript quantification was also verified with RT-PCR (Figure 3).
Why I like this preprint
While various tools are already in use for the manipulation of gene expression, there are still drawbacks to existing techniques as the CRISPR-Cas9 system and RNAi. For example, the strategies in the RNAi method currently include using larger dsRNAs to trigger the RNAi machinery as a means to overcome the problem of unpredictability of siRNA sequences generated. This approach increases the probability of off-target collateral effects. Other studies carried out on Cas13a have not demonstrated the same type of non-specific promiscuity, although this remains contentious and further studies need to be carried out. Nonetheless, increasing the repertoire of gene-editing tools allows for higher specificity in these techniques, especially when different organisms are used.
Questions
- In addition to RNA cleavage, have there been any studies that have exploited CRISPR-Cas13 for insertions? If so, how do these compare or differ to the Cas9 system?
- In terms of its crRNA sequence, how does Cas13 mechanistically target mRNA instead of DNA compared to Cas9? Does this have to do with the lack of the PAM?
References
- Tijsterman M., Plasterk R., 2004. Dicers at RISC: The Mechanism of RNAi, Cell, 117 (1).
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F., 2013. Genome engineering using the CRISPR-Cas9 system, Nature Protocols, 8 (2281-2308).
- Marraffini L.A., Sontheimer E.J., 2008. CRISPR interferences limits horizontal gene transfer in staphylococci by targeting DNA. Science, 322 (1843-1845).
- Clements M., Boocock R., 2008. Ovarian development in mosquitos: Stages of growth and arrest, and follicular resorption, Physiology Entomology, 9 (1-8).
- Isoe J., Collins J., Badgandi H., Day W.A., Miesfield R.L., 2011. Defects in coatomer protein I (COPI) transport cause blood feeding-induced mortality in Yellow Fever mosquitoes, Proc Natl Acad Sci USA, 108 (211-217).
doi: https://doi.org/10.1242/prelights.15734
Read preprint










 (No Ratings Yet)
(No Ratings Yet)