SUV39 SET domains mediate crosstalk of heterochromatic histone marks
Posted on: 3 July 2020 , updated on: 8 July 2020
Preprint posted on 2 July 2020
Article now published in eLife at http://dx.doi.org/10.7554/elife.62682
Heterochromatin establishment relies on cross-talks between H3K9 methylation and H3K14 ubiquitylation
Selected by Yasmin LauBackground
In both unicellular and multicellular organisms, the maintenance and transmission of certain epigenetic states plays a vital role in controlling gene transcription spatially and temporally. In addition to its implications in disease, epigenetic activity is also essential for many cellular processes and can be influenced by environmental cues, thus providing adaptational value (2). Such states can be achieved by a concerted mechanism between DNA methylation, histone modifications and microRNA activity, which determines chromatin structure (2). The heterochromatic mode of chromatin is typically characterized by transcriptional repression and dense packing mediated by histone modifications (1). Specifically, di- and trimethylation of the lysine 9 on histone 3 (H3K9me2/3) is a highly conserved feature of heterochromatin establishment, which is integral for maintaining genome stability and the timely regulation of gene expression programs (1).
Many aspects of the mechanisms underlying heterochromatin establishment still remains poorly understood. Recent studies have revealed that the protein Clr4 in the fission yeast Schizosaccharomyces pombe, a lysine methyltransferase containing a SET-domain in the SUV39 clade, functions to deposit the the H3K9me mark across the yeast genome (3). Akin to many of its SUV39 orthologs in higher organisms, Clr4 contains an N-terminal chromodomain and a C-terminal SET catalytic domain (Fig 1) which mediates binding to H3K9 and confers methyltransferase activity respectively (4). Transcriptional gene silencing through heterochromatin formation relies on an RNA interference mechanism which involves both the recruitment of Clr4 as well as Cullin4-RING ubiquitin ligase complex (the CLRC complex), which has been revealed to also underlie ubiquitylation of H3K14, another hallmark of heterochromatin (3).
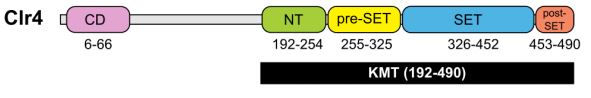
Figure 1: The CD and KMT domains of Clr4
Previously, Clr4 activity has been shown to be stimulated by H3K14ub by the CLRC complex (3). In this preprint, the authors further characterized the crosstalk between H3K14 ubiquitination and Clr4 activity and the role of this on heterochromatin function in S. pombe.
Key Findings
The authors first sought to understand the subunits of the Clr4 architecture involved in stimulation by H3K14ub. This was achieved by carrying out measurements of methyltransferase activity in both full length Clr4 and the KMT domain alone using H3K14ub and H3K18ub as substrates. Compared to full length Clr4, the isolated KMT domain showed a higher stimulation by H3K14ub, and H3K18ub gave rise to 40-50 fold lower stimulation for both constructs which highlights the specificity of Clr4 to lysine 14. The authors also carried out a separate enzyme kinetics assay focusing on Clr4KMT activity with H3K14ub and unmodified H3, where a hundred-fold increase in Michaelis-Menten constants was seen in H3K14ub compared to H3. Thus, the effect of H3K14ub on Clr4 points to the KMT domain which has a high binding affinity for the ubiquitylated H3K14.
Next, the structural regulation of Clr4 was investigated using hydrogen/deuterium exchange (HDX) mass spectrometry. The HDX rates for free Clr4-KMT and Clr4-KMT with H3K14ub or H3 were measured, which reflects the regions of the protein that are affected by binding. A sharp change in HDX was observed for Cl4-KMT with H3K14ub between residues 243-261, while residues 291-305 showed amide protection in both H3K14ub and H3 complexes. Interestingly, these residues correspond with the “back” of the Clr4 structure, where the opposite side comprises of pockets for cofactor and substrate peptide binding (Fig 2).
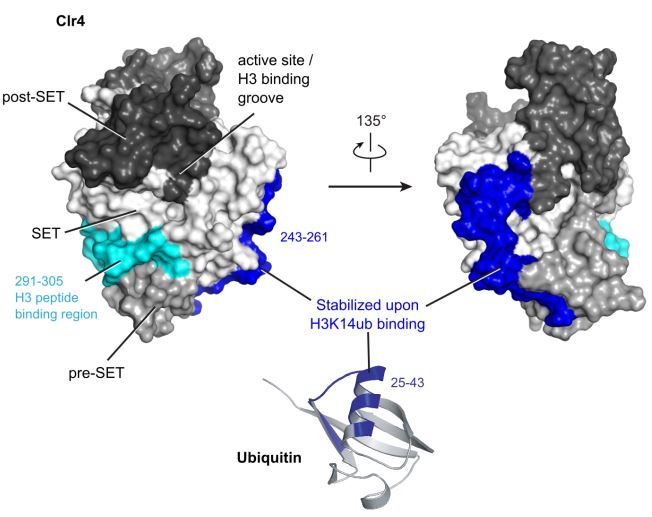
Figure 2: Scheme of Clr4 and ubiquitin structures
To decipher the role of the residues within the H3K14ub region that displayed the highest HDX rate, loss-of-function mutants were obtained and assayed with ITC (isothermal titration calorimetry). Amongst this region, replacement of residues 253-256 (Clr4GS253) yielded significant loss of binding to H3K14ub. This loss of affinity was also seen upon 3 phenylalanine substitutions close to this region (F253A, F310A and F427A). These mutants were also subjected to methyltransferase activity testing with H3K14ub and H3. All mutants displayed a loss of activity with H3K14ub compared to the wild-type Clr4-KMT; however, loss of activity was only observed for Clr4GS253 with H3, while F3A mutants were similar to the wild-type. This suggests that stimulation of Clr4 is specific to ubiquitinated H3K14 and this cross-talk relies on the 253-256 stretch of amino acids.
Finally, the authors utilized the Clr4GS253 and F3A mutants to probe their importance in Clr4 stimulation by H3K14ub and heterochromatin function. Transcriptional gene silencing was first qualitatively assessed with growth phenotypes on 5-FOA with mutants containing a ura4-associated centromeric reporter, such that wild-type strains with non-defective silencing repress this transcript and grow poorly on media lacking uracil. Similar to the clr4Δ mutant, Clr4GS253 exhibited a significant loss in transcriptional gene silencing, while a similar but milder phenotype was seen for F3A mutants. Transcriptional gene silencing in these mutants was also quantified by measuring levels of heterochromatic transcripts, which were highly elevated in both clr4Δ and Clr4GS253 and modestly so in F3A. Thus, albeit at different severities, these mutants exhibit silencing defects induced by impaired Clr4 activity.
Why I like this preprint
The aspect of this preprint that I like the most is that a wide variety of biochemical, chemical as well as genetic techniques have been used to validate these findings, ranging from qualitative growth assays to HDX mass spectrometry. Such findings also highlight specific features of epigenetic mechanisms which entail cascade cross-talk stimulation of one modification (H3K9me) by another (H3K14ub), and that these features are also conserved from single cell organisms such as fission yeast to mammals.
Questions
- In the process of selecting mutants for the functional assays, what features of the 3 phenylalanine residues chosen made them conspicuous for study? Is it simply that they are in close proximity to the Clr4GS253 region or are there also other factors?
- Is it known whether other SUV39 SET-domain proteins like Clr4 are also stimulated by specific modifications such as ubiquitylation?
References
- Peng, J. C., & Karpen, G. H. (2007). H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nature cell biology, 9(1), 25–35.
- Xu, W. et al. (2016). Epigenetics and Cellular Metabolism. Genetics & epigenetics, 8, 43–51.
- Oya, E. et al. (2019) H3K14 ubiquitylation promotes H3K9 methylation for heterochromatin assembly. EMBO Rep. 20, e48111.
- Akoury, E. et al. (2019) Disordered region of H3K9 methyltransferase Clr4 binds the nucleosome and contributes to its activity. Nucleic acids research, 47(13), 6726-6736.
doi: https://doi.org/10.1242/prelights.22602
Read preprint










 (No Ratings Yet)
(No Ratings Yet)
5 years
Alessandro Stirpe
Dear Yasmin,
I am really happy that you enjoyed our preprint!
To answer your questions:
1. The 3 phenylalanine residues were chosen in part because close to the region identified by HDX mass spec, and in part because ubiquitin-binding motifs tend to often interact with a hydrophobic patch present on Ubiquitin.
2. We tested various SUV39 proteins: the human proteins G9a and SUV39h2, and the plant SUVH4. Among these proteins, SUV39h2 is stimulated by H3K14ub and shows a behaviour similar to Clr4, indicating a likely conservation for the mechanism we described.
Best,
Ale