Preprints by preLighters: Clarice Hong
16 March 2021
Clarice Hong is a graduate student in Barak Cohen’s lab at Washington University in St Louis, where she studies how cis-regulatory sequences work together to control gene expression. She has been an active preLighter since mid-2018, and has written ten preprint highlights so far covering various aspects of gene regulation. Clarice recently posted her first preprint from her PhD work:
Genomic environments scale the activities of diverse core promoters
Clarice KY Hong, Barak A Cohen
doi: https://doi.org/10.1101/2021.03.08.434469
We asked Clarice to tell us more about her discoveries and the reason they shared the study on bioRxiv.

Could you explain the main findings of your work?
Cell-type specific gene regulation requires precise spatiotemporal coordination between core promoters and their surrounding enhancers and chromatin environments. Classical studies of position effect have shown that the surrounding genomic environments have large effects on gene expression. However, it is less clear whether the effects are the same on every core promoter, or whether they are specific to particular classes of core promoters.
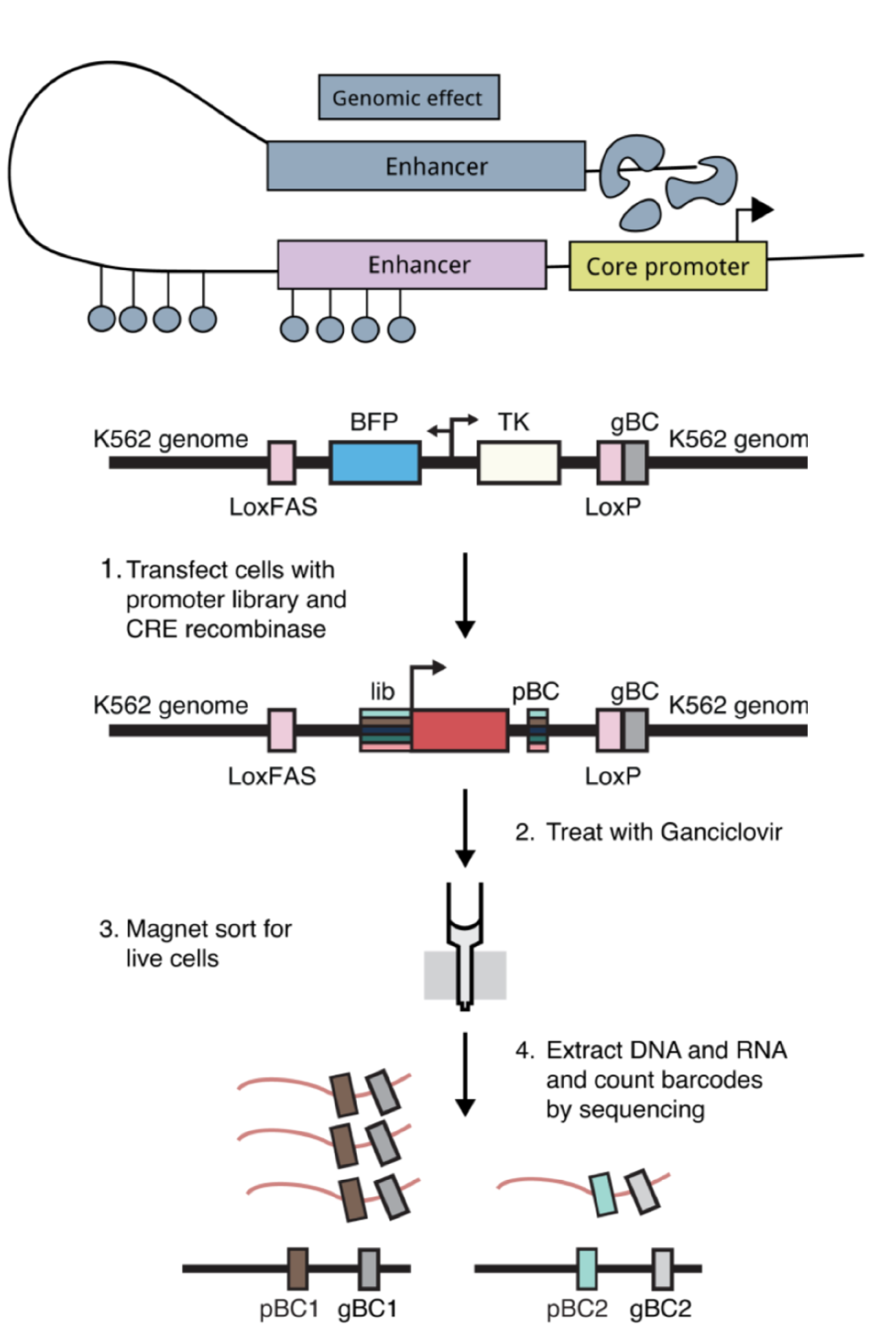
One model for how genomic environments interact with core promoters is that core promoters with different sequence elements respond specifically to distinct genomic environments containing different enhancers and chromatin features. For example, housekeeping and developmental core promoters have been shown to respond to distinct classes of enhancers and respond to distinct sets of proteins1–3. Core promoters containing either TATA or DPE motifs in the same genomic location in Drosophila have also been shown have different expression patterns because they respond to different enhancers4. To test this model, we measured the activity of hundreds of diverse core promoters at four locations in the genome using a genome-integrated massively parallel reporter assay (MPRA) that our lab had previously developed5. As a complementary experiment, we also measured the activity of six selected core promoters (three housekeeping and three developmental) in thousands of locations across the genome using the Thousands of Reporters Integrated in Parallel (TRIP) assay6.
Intriguingly, we found that while average expression of core promoters was heavily dependent on their genomic location, the relative strengths of the core promoters were large preserved across genomic locations. Housekeeping and developmental promoters responded to genomic environments in the same way, suggesting that different promoter classes do not interact specifically with genomic environments. We also measured the intrinsic activities of the core promoters on plasmids and found that the intrinsic activities correlate well with their activities in the genome. This suggests that the main role of diverse promoter motifs is to set the strength of the core promoters, which are then scaled up or down according to the surrounding genomic environment.
Finally, even though the rank order of core promoters is preserved across locations, the quantitative differences between promoters can be different. We show that this is because promoters of different intrinsic strengths respond to changes in genomic environments in different ways. Strong promoters are able to overcome less active genomic environments, while weaker promoters require more active genomic environments for high expression. Thus, promoter strength, not class, determines the quantitative extent to which it responds to genomic environments.
Our work dramatically simplifies the current working model in which promoters achieve specificity, which is thought to be through extensive and specific interactions with the genomic environments. Instead, our results argue for a modular genome lacking sequence-specific interactions between core promoters and their genomic environments.
Why did you decide to post your work on bioRxiv?
I believe that research is moving faster than the current publishing system. If we all waited for our papers to be published before releasing it to the community it would greatly slow down the pace of scientific discoveries. In fact, there was a recent preprint that had results that really agreed with what we found, it would have taken months for us to find out if we waited for the publication of the paper. I also think it is a great way to get feedback from the community quickly – after working on the project for so long I can no longer determine how interesting or useful my work is, but the response that I have seen on Twitter in the few days since it’s been out has been super encouraging for me! Lastly, I feel strongly about making research accessible and available to everyone, and bioRxiv is open-access and free for all 😊 I would encourage everyone to preprint their papers if possible!
References
- Zabidi, M. A. et al. Enhancer–core-promoter specificity separates developmental and housekeeping gene regulation. Nature 518, 556–559 (2015).
- Arnold, C. D. et al. Genome-wide assessment of sequence-intrinsic enhancer responsiveness at single-base-pair resolution. Nat. Biotechnol. 35, 136–144 (2017).
- Haberle, V. et al. Transcriptional cofactors display specificity for distinct types of core promoters. Nature 570, 122–126 (2019).
- Butler, J. E. F. & Kadonaga, J. T. Enhancer–promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 15, 2515–2519 (2001).
- Maricque, B. B., Chaudhari, H. G. & Cohen, B. A. A massively parallel reporter assay dissects the influence of chromatin structure on cis-regulatory activity. Nat. Biotechnol. 37, 90–95 (2019).
- Akhtar, W. et al. Chromatin Position Effects Assayed by Thousands of Reporters Integrated in Parallel. Cell 154, 914–927 (2013).










