Meet the preLighters: an interview with Tessa Sinnige
23 May 2019
Tessa Sinnige is a postdoctoral fellow and she recently moved to Northwestern University, USA from the University of Cambridge. In her research she studies the mechanisms of protein aggregation using C. elegans as a model. We caught up with Tessa to discuss about her science, postdoc life, preprints and her experience with preLights.
What got you into science, and biochemistry in particular?
I was always fascinated by the human body, how it works and what goes wrong in disease. And I always liked going into the details. So I guess that’s why I went into the direction of structural biology and biophysics; to look at protein molecules and how they were moving and interacting with each other. I did my Master’s at Utrecht University, which is really strong in structural biology, and got to do two research projects. I remember being really excited to go into the lab, rather than just taking courses.
And during your PhD, what topic were you working on?
I really wanted to work on amyloid fibrils, because I got very interested in protein folding, misfolding and aggregation. At that time solid-state NMR was the best technique to determine the structures of amyloid fibrils, so I joined the lab of Marc Baldus, hoping that I could work on them. But it doesn’t always work out the way you plan it. I ended up working on a membrane-protein complex that is responsible for folding other membrane proteins in the outer membrane of Gram negative bacteria. So at least it was still related to protein folding, and it was a cool and challenging project which really pushed the limits of solid-state NMR.
For your postdoc you switched from structural biology to imaging in C. elegans. Could you tell us more about this decision and the reason behind it?
It took me a while to make the decision, because this time I was quite determined to work on protein aggregation. I actually visited 8 different labs before deciding where to do my postdoc.
I also wanted to make the change to a biological model system, to increase the biological relevance of the research I was doing. Talking to a lot of people and visiting all those labs, I came across C. elegans. I thought the worm is an interesting experimental system to look at age-related protein aggregation, because C. elegans is also ageing – in a similar way that humans do – but you can do things on a relatively short time-scale. You can directly see the aggregates that are formed by fluorescently labelled proteins, and you also have good readouts for the related toxicity, such as changes in the lifespan and the behaviour of the worm. So I decided to join the Centre for Misfolding Diseases in Cambridge, which is originally a biophysics lab, but they were very excited about using C. elegans as a model organism.
What is the biggest question you would like to answer with your research?
I basically want to know how proteins aggregate in vivo. Biophysical studies on protein aggregation have largely been done on purified proteins, so this is where most of our knowledge about the kinetics and the structural aspects comes from. But in vivo we don’t really know if the mechanisms are the same, and the time scales are definitely different. In humans it can take decades for proteins to aggregate and to cause damage.
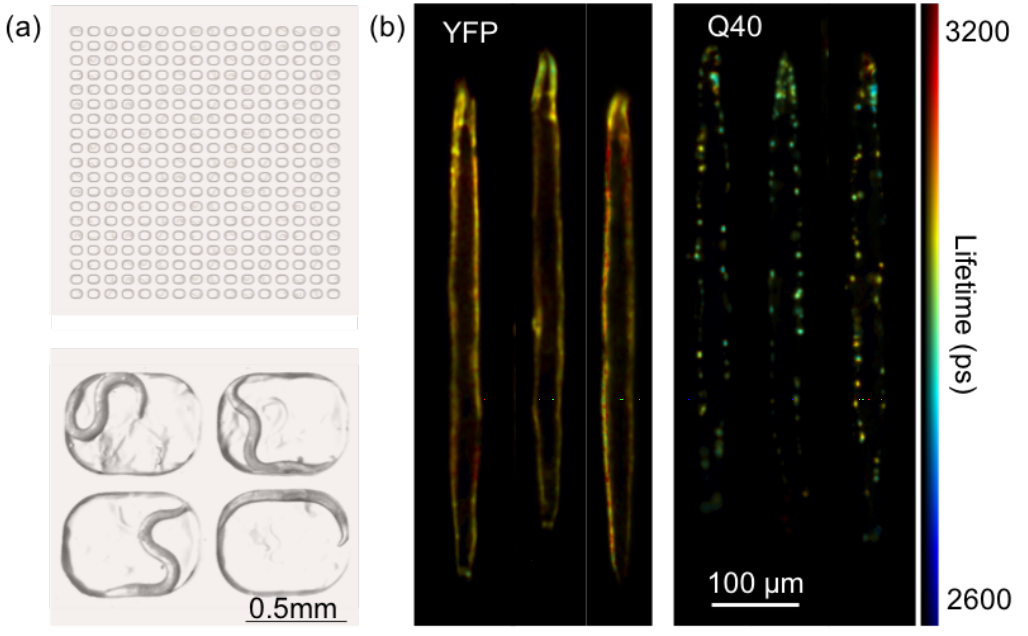
Because performing molecular level studies in vivo is technically very challenging, a large part of my postdoc was about setting up methods that would allow me to do this. What was really nice is that I collaborated with the labs of Clemens and Gabi Kaminski; one of their postdocs had built a fluorescence lifetime imaging microscope that allowed us to image amyloid formation in a population of worms and follow this as they were ageing. We found some really interesting differences between proteins; a protein related to Huntington’s disease aggregated straight away when the worms were still young, but a protein linked to Parkinson’s disease was actually forming little blobs that weren’t the typical amyloid-like aggregates, until the worms were very old. So it’s a totally different aggregation mechanism. You can actually read more about this story on bioRxiv, and I’m happy to share that the paper just got accepted!
Why did you decide to preprint this work? And in general, what’s your take on preprints?
It took quite a long time to decide where to submit and to get everything ready, since it was a pretty large collaboration with many people involved. I started applying for a new position and wanted some evidence that this work had been done, so that’s one of the reasons we decided to post a preprint. It’s been great that I could mention it in the job applications and talks that I’ve been giving. It’s much more concrete to have a preprint than saying ‘manuscript in preparation’ or ‘manuscript submitted’.
One thing that I really like about preprints is that manuscripts get appreciated for their scientific value, and not for some crazy impact factor. That’s what I also like about preLights; that we select the preprints that we think are interesting for the community.
On the other hand, I want to mention that we didn’t really receive feedback on our preprint, so that aspect of pre-printing probably still needs some more time to develop. And of course that’s also where preLights comes into the picture.
So what was your main motivation for joining preLights?
One reason was that I really like writing and I thought I could gain new skills, because writing a preLight is quite different from writing a paper or proposal. And I think it really worked out, because I got a better feeling for writing for a slightly broader audience, becoming more creative and giving my posts a bit of a personal touch. Another motivation was to keep an eye on the preprints that are coming out, and I also found that you have to read them in much more detail than you normally would if you decide to write a preLight. And the third reason was that I was interested to get involved in peer-reviewing, but I never got asked by anyone! Even though writing preLights is slightly different, it’s certainly a step in this direction, and I thought that it might even get picked up by editors. It would make sense to get postdocs involved in peer-review as independent people, not just having them review for their supervisor – I actually just highlighted a preprint about this topic together with Gautam Dey.
And what’s been your experience being a preLighter so far?
One of the things I find most rewarding is when I get a response from the preprint authors about the questions I raised. Then I’m always really happy that I made the effort to write the preLight. And I think the response from the authors also gives a lot more depth to the story, because you get to see more about why they actually did the study and how they are thinking about following up.
Also, the interaction with the other preLighters has been a positive surprise to me. It’s a really nice community and the communication via Slack is very convenient. I discovered a few people who are interested in similar things, and you end up reading each other’s preLights and giving feedback. And there are always some discussions happening for example about publishing or other things, so I’m happy to be part of that.
“The interaction with the other preLighters has been a positive surprise to me […], I discovered a few people who are interested in similar things, and you end up reading each other’s preLights and giving feedback.”
Getting back to your postdoc, you have been involved in quite many social activities, could you tell us more about it?
Cambridge is a great place to be a postdoc, there are over 4000 of us and it’s a very lively community. The postdoc society organizes a lot of events, some professional but also some social. I really enjoyed participating in these events, and then after some point I also started organizing them – because of course you need people to make these things happen. I co-organized a couple of hiking trips where we explored a lot of beautiful nature in the UK. So for me this was a nice way to get to know the country. I also organized a trip to London to see a ballet, and some other events like going to a jazz concert or having a wine and cheese night. It was great because you meet people from different fields and from all over the world, which is fun and you learn a lot from it.
What are your plans for the near future?
Right now I’m very excited about starting a new adventure in the US in the Morimoto lab, it was the perfect opportunity to continue a project that I’ve started in Cambridge. But in the long term I would like to be in Europe, so I’m mainly looking for group leader positions there.
Is there anything people would be surprised to find out about you?
I’m really an open book, so I don’t think there is much secret about me. I love jazz, I used to play the flute and the saxophone, but now I rather go to concerts and listen to people who really know how to play. I like art and museums and other cultural things as well, and I also like travelling which is a good thing as a scientist.










