Preprints by preLighters – Teresa Rayon
20 April 2021
Teresa Rayon is a postdoctoral researcher in James Briscoe’s lab at the Francis Crick Institute in London, where her research focuses on stem cells and embryonic mammalian development. She has been a preLighter since the very beginning of preLights and has written a number of preprint highlights during that time.
Teresa recently posted a preprint from her postdoc, so we caught up with her to ask her about her new research, why she wanted to post it as a preprint, and what’s next for her. If you’d like to find out more about Teresa, you can read an interview we did with her back in 2019 here and follow her on Twitter @t_rayon
Single cell transcriptome profiling of the human developing spinal cord reveals a conserved genetic programme with human specific features
Teresa Rayon, Rory J. Maizels, Christopher Barrington, James Briscoe
bioRxiv (2021) https://doi.org/10.1101/2021.04.12.439474

Could you tell us the main findings in your preprint?
The patterning and differentiation of the developing spinal cord is one of the most beautiful and better studied developmental processes across species (Briscoe et al., 2015; Davis-Dusenbery et al., 2014; Le Dréau et al., 2012; Tao and Zhang, 2016; Zhang et al., 2010). However, the molecular understanding of spinal cord development in human embryos remains largely limited (Betters et al., 2010; Bondurand et al., 1998; Dady et al., 2021; Marklund et al., 2014; O’Rahilly and Müller, 2007). This is mostly due to the difficulty of access to the material, as the tissue needs to be donated voluntarily by women who have had a termination of pregnancy (abortion) and collected following strict regulation. With the support from the Wellcome Trust-MRC Human Developmental Biology Resource (https://www.hdbr.org/general-information) in the UK, we collected the spinal cord of four human embryo stages that span from gestational week (W) 4 to W7 of pregnancy and profiled the transcriptomes of the cells in the tissue.
In the preprint, we have identified spinal cord and peripheral neural cell types in human embryos, compared differences and similarities of progenitor patterning in human and mouse, inferred the dynamics of neurogenesis within the neural tube, compared the dataset with in vitro human stem cell motor neuron differentiations, and generated an online resource for the community.
I would highlight the overall conservation of genetic programs between mouse and human, and the limited yet exciting human-specific features that we identified. In addition, I’m looking forward to seeing if the dataset is used as a resource in the community to study the embryonic nervous system and whether stem cell biologists make use of the in vivo data to benchmark in vitro models of human neural development.
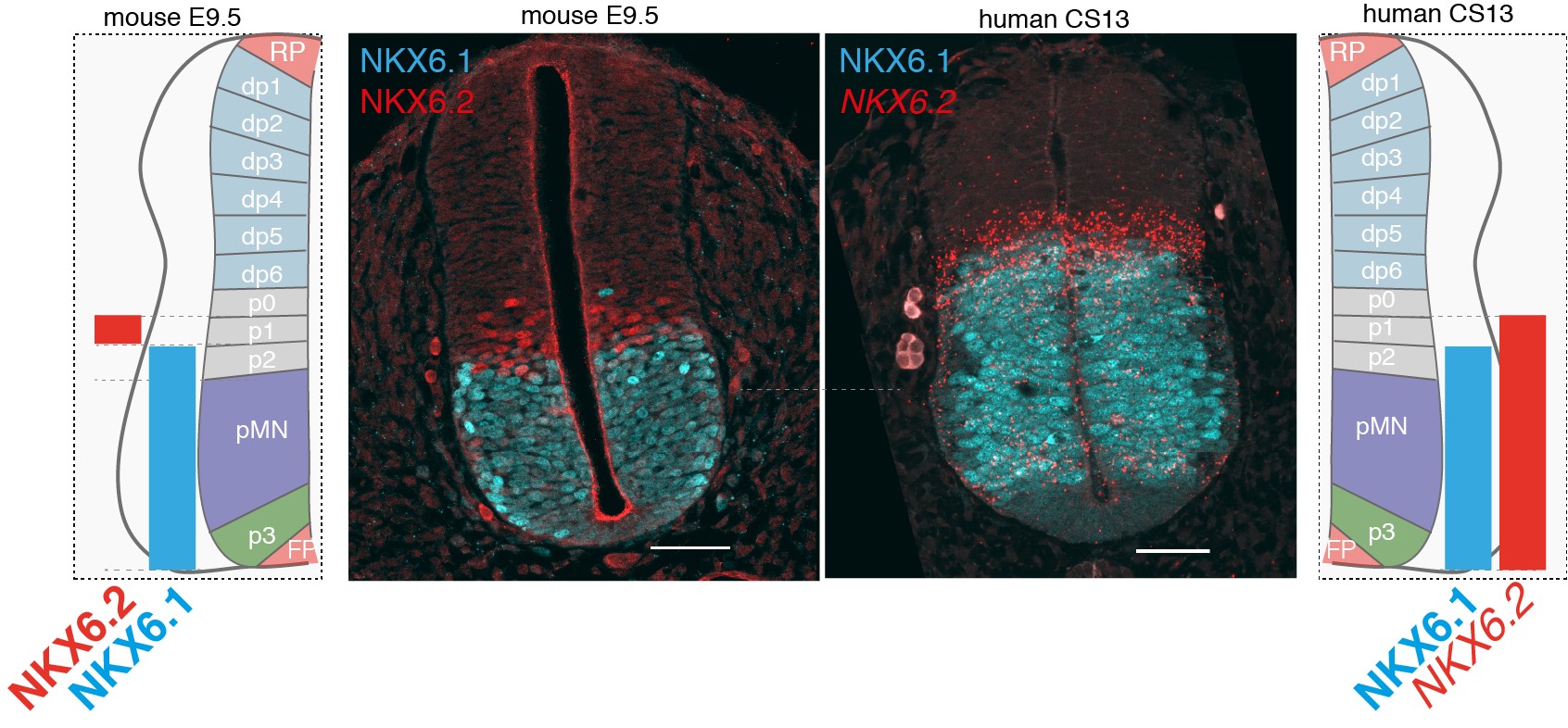
Differences in NKX6.2 expression in the developing mouse and human spinal cord. NKX6.1(cyan) and NKX6.2 (red) expression in transverse sections of the mouse and human neural tube at shoulder levels in E9.5 and CS13. Scale bars, 50 microns (um).
Why did you decide to preprint your work?
This project has been a long time in the making. Everything started in 2018 when we designed the experiments for the mouse neural tube atlas (Delile et al., 2019). We collected the human embryos between October 2019 and January 2020, and the analysis has taken place during the COVID pandemic. As science benefits from the rapid and efficient dissemination of results and the availability of the human material is very restricted to researchers, we thought that posting the work on bioRxiv was the best approach.
To me, preprints allow you to publish your work when you feel it is ready to be shared. This makes it exciting as the data is still fresh and the feedback from others can still be integrated in the work, prior to peer-review publishing. Moreover, I’m now in the job market applying for group leader positions. The preprint allows me to show what I’ve been working on, and evaluators can read and judge the work for themselves.
What’s next for you?
My plan is to start an independent research group studying how timing is encoded during development. I’ve spent the last year applying for grants, submitting job applications and performing zoom-interviews while writing the manuscript. Hopefully, I will be able to start my lab soon!
REFERENCES
Betters, E., Liu, Y., Kjaeldgaard, A., Sundström, E. and García-Castro, M. I. (2010). Analysis of early human neural crest development. Developmental Biology 344, 578–592.
Bondurand, N., Kobetz, A., Pingault, V., Lemort, N., Encha-Razavi, F., Couly, G., Goerich, D. E., Wegner, M., Abitbol, M. and Goossens, M. (1998). Expression of the SOX10 gene during human development. FEBS Letters 432, 168–172.
Briscoe, J., Small, S., Butler, S. J., Bronner, M. E., Kicheva, A., Bollenbach, T., Ribeiro, A., Valle, H. P., Lovell-Badge, R., Episkopou, V., et al. (2015). Morphogen rules: design principles of gradient-mediated embryo patterning. Development 142, 3996–4009.
Dady, A., Davidson, L., Halley, P. A. and Storey, K. G. (2021). Human spinal cord differentiation proceeds rapidly in. bioRxiv 2021.02.06.429972.
Davis-Dusenbery, B. N., Williams, L. A., Klim, J. R. and Eggan, K. C. (2014). How to make spinal motor neurons. Development 141, 491–501.
Delile, J., Rayon, T., Melchionda, M., Edwards, A., Briscoe, J. and Sagner, A. (2019). Single cell transcriptomics reveals spatial and temporal dynamics of gene expression in the developing mouse spinal cord. Development 146, dev173807.
Le Dréau, G., Martí, E., Le, G., Au, D. and Martí, E. (2012). Dorsal-ventral patterning of the neural tube: A tale of three signals. Developmental Neurobiology 72, 1471–1481.
Marklund, U., Alekseenko, Z., Andersson, E., Falci, S., Westgren, M., Perlmann, T., Graham, A., Sundström, E. and Ericson, J. (2014). Detailed Expression Analysis of Regulatory Genes in the Early Developing Human Neural Tube. Stem cells and development 23, 5–15.
O’Rahilly, R. and Müller, F. (2007). The development of the neural crest in the human. Journal of Anatomy 211, 335–351.
Tao, Y. and Zhang, S.-C. (2016). Neural Subtype Specification from Human Pluripotent Stem Cells. Cell Stem Cell 19, 573–586.
Zhang, X., Huang, C. T., Chen, J., Pankratz, M. T., Xi, J., Li, J., Yang, Y., LaVaute, T. M., Li, X.-J., Ayala, M., et al. (2010). Pax6 Is a Human Neuroectoderm Cell Fate Determinant. Cell Stem Cell 7, 90–100.










