Preprints by preLighters – Tessa Sinnige
15 September 2020
Tessa Sinnige has been part of the preLights team for nearly two years and has highlighted a number of preprints related to protein aggregation (her main research focus), but also studies on other topics such as academic research culture (you can read all her preLights here). Following the ‘Meet the preLighters’ interview last year, we asked Tessa about her latest preprint and the research plans for the new lab she started this September.
Kinetic analysis reveals the rates and mechanisms of protein aggregation in a multicellular organism
Sinnige et al. (2020) bioRxiv doi.org/10.1101/2020.08.13.249862
Could you explain the main findings of your new preprint?
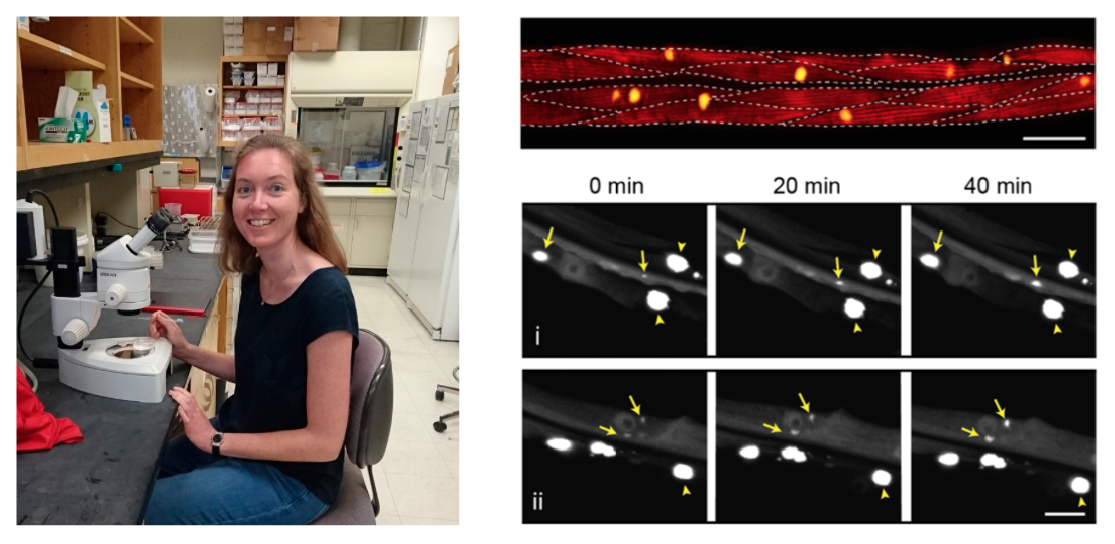
Protein aggregates containing amyloid fibrils are found in many different diseases, so there is a lot of interest in understanding how these structures form. However, most of our current knowledge comes from test tube reactions with purified proteins. In these systems, it is possible to systematically analyse the kinetics of amyloid formation and derive the microscopic reaction steps and rates by mathematical modelling. In our preprint, we apply these methods to a living animal for the first time using a C. elegans model expressing polyglutamine in the muscle cells. Our experimental data point to a mechanism of stochastic nucleation, in which aggregates form at different moments in time in each of the muscle cells, consistent with what we know about the biophysics of protein aggregation in small volumes. Using mathematical modelling we found that the overall nucleation rate is however constant, and that the cells are not influencing each other’s aggregation propensity but operate as individual entities within the animal. We also found that aggregate growth is really fast once nucleation has occurred, and this is why most cells acquire only one big aggregate. The same has been previously observed for polyglutamine aggregates in patient material and mouse models, so our analysis may be relevant to human disease.
Why did you decide to post your work on bioRxiv?
One of the main advantages of preprint servers such as bioRxiv is that the manuscript is available straight-away, whereas it can easily take another year before it would end up in a journal. This is of course important for science as a whole, but also for the careers of individual researchers. In my case, my preprints have been very important for job and grant applications.
You are just starting up your lab, what will be the main focus of the Sinnige lab (at least initially)?
I am really building on this latest preprint, hence its importance! Now that we have a mechanism for polyglutamine aggregation in C. elegans, we can move on to some very interesting biological questions and for example look at the role of molecular chaperones. We now also have the tools to explore how different tissues and cell types respond to protein aggregation, and to compare different disease-associated proteins, to learn more about the rules that govern protein aggregation in vivo.