Decoding commensal-host communication through genetic engineering of Staphylococcus epidermidis
Posted on: 2 July 2019
Preprint posted on 10 June 2019
Getting Under Staphylococcus’s Skin – Genetic Manipulation of S. epidermidis Reveals the Molecular Basis of Host-Microbe Interactions
Selected by Connor RosenBackground:
The immune system is shaped by microbial exposure at all barrier surfaces. This includes the commensal microbiota, which colonizes without gross barrier disruption or inflammation, and is necessary for healthy immune maturation. One dominant member of the human skin microbiome is Staphylococcus epidermidis, which acts as a commensal to induce non-pathogenic immune responses important for normal skin homeostasis. S. epidermidis can also act as an opportunistic pathogen, causing disease upon systemic dissemination or loss of regular immune function. Precise understanding of the interactions between S. epidermidis and the immune system in settings of both health and disease is limited due to the genetic intractability of many S. epidermidis isolates. Here, Chen et al develop a broadly applicable genetic system for S. epidermidis and use this to explore the bacterial factors involved in commensalism and infection.
Key findings:
- Development of genetic tools for S. epidermidis strains
The authors make tremendous strides enabling the broad genetic manipulation of S. epidermidis. Key steps included the use of methylase-deficient E. coli strains for donor DNA production, hyperosmolar sorbitol outgrowth, extensive glycerol washes, and strain-specific timing of heat shocks before or after electroporation. These resulted in manipulation of 13 distinct isolates of S. epidermidis (of 17 tested).
- PRR engagement drives differential CD8 and CD4 responses
The authors colonized mice deficient in various pattern recognition receptors (PRRs) to study the impact of microbial sensing on the skin immune response, focusing on TLRs and the C-type lectin receptor Dectin-1. Examination of the adaptive responses (CD4 and CD8 T cells) revealed that CD4 and CD8 numbers, as well as polarization towards IL-17 or IFN-g production, were not uniformly influenced by individual PRRs, but different knockouts selectively impacted distinct immune cell subsets. Using the genetic tools they developed, they also colonized mice with S. epidermidis deficient in various cell wall components that are ligands for PRRs, and observed that different components were key for stimulating distinct responses. Together, these results all suggest that distinct sensory modules, stimulated by individual microbial ligands, are responsible for different components of the adaptive immune response to commensals, rather than pan-microbial responses being equally activated.
- Invasion and colonization drive distinct immune reactions
Finally, the authors examined whether the distinct microbial ligand roles identified in colonization held true during infection models with the same bacteria. While infection with various cell-wall component deficient S. epidermidis strains led to approximately equivalent adaptive immune responses, the innate immune responses (e.g. neutrophil infiltration) varied more with different mutants. This suggests that distinct microbial ligands drive different responses in both infection and colonization, but the precise role for particular ligands may be different between each setting.
Importance:
This preprint firmly establishes a genetic system for S. epidermidis manipulation, which will be key to advancing studies of this microbe and the skin microbiome in general. Additionally, it helps establish a paradigm for the distinct sensory roles of various PRRs, the different cell types expressing them, and their respective microbial ligands, in establishing healthy commensalism and homeostatic immune responses. These observations may serve as a general starting point to understand complex immune responses to individual microbes and how those responses are altered during opportunistic infections or manipulated by “pathobionts”. Finally, this work represents a strong model for molecular understanding of host-microbe interactions, using both host and microbial genetics to interrogate the immune response to a microbe and the precise microbial factors involved in driving that response (Figure 1).
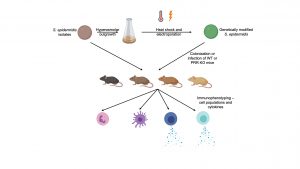
Figure 1. Human S. epidermidis isolates can be genetically manipulated using an optimized protocol involving hyperosmolar culture in sorbitol and heat shock combined with electroporation. Colonization of WT or PRR KO mice with parental or genetically modified S. epidermidis, with subsequent immunophenotyping, reveals the roles of different microbial ligands and their sensors in driving different facets of the immune response. Figure prepared with Biorender.
Moving Forward:
- The authors do note a difference in T cell infiltration (both CD4 and CD8) in TLR5 deficient mice. Staphylococci are generally described as non-motile and without flagellin, the ligand for TLR5. Does S. epidermidis express flagellin, or is there a more complicated regulation at play (perhaps involving indirect regulation of sensation of other skin microbes which may express flagellin)?
- What scale of transformation efficiencies are achieved using this new genetic method, and how variable is this between strains? Is it amenable to genome-scale efforts such as introduction of transposon libraries?
- The difference between colonization and infection is very interesting and could be explained by several models. For example, is the key difference the ability of non-myeloid cells in the skin to directly sense S. epidermidis during colonization, but not infection? Or do the differences rely on immune “education” in the skin – does the presence of the normal flora “train” the skin-resident immune cells towards a commensal-favoring phenotype, while the circulating immune cells responding to infection have not received the same prior signals? Would colonization of germ-free mice, with no prior microbial exposure, more closely mimic SPF infection or colonization?
- The authors show that for at least Dectin-1, a non-myeloid (radio-resistant) population is responding to S. epidermidis. It will be intriguing to see whether the effects of this are cell-intrinsic (impacting those cells and their response to S. epidermidis alone), or function through “cross-talk” with immune cells.
doi: https://doi.org/10.1242/prelights.11683
Read preprint










 (No Ratings Yet)
(No Ratings Yet)