Evidence that the cell cycle is a series of uncoupled, memoryless phases
Posted on: 27 April 2018
Preprint posted on 16 March 2018
Article now published in Molecular Systems Biology at http://dx.doi.org/10.15252/msb.20188604
Forget and move on: phase durations in the unperturbed mammalian cell cycle are uncorrelated
Selected by Dey LabCo-authored with Agathe Chaigne, Sir Henry Wellcome Fellow at the MRC LMCB
Context
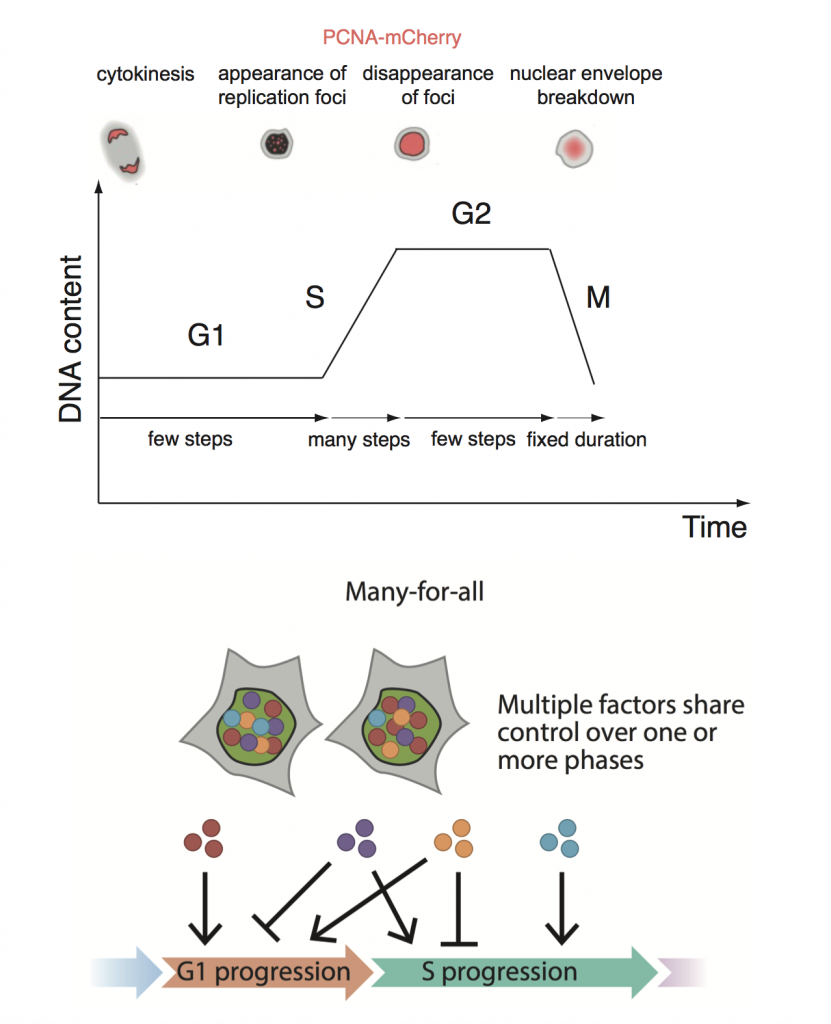
The metazoan cell cycle is canonically defined by DNA replication (S) and cell division (M) periods that are delimited by G1 and G2 “gap” phases. Despite a wealth of research accumulated over several decades on the regulatory circuits and checkpoints that govern each phase transition, direct measurements of cell cycle variability awaited the recent development of live cell reporters1,2.
The following data in the literature set the tone for the Chao et al. preprint: variability in cell cycle duration can stem from either G13 or S/G24, M phase is temporally insulated from the rest of the cell cycle5, and phase durations in sister cells are correlated6. The authors set out to investigate the question of phase coupling in more detail, using a PCNA fluorescent reporter in three human cell lines.
Key findings
Despite pronounced variability between cell types and within individual cell lines, the authors found no correlation between phase durations in individual cells in any of their cell types. At the same time, and consistent with the literature, phase durations in sister cells were strongly correlated with each other. Moreover, none of a set of physicochemical perturbations to G1, S or G2 were able to induce any additional correlation between phases.
The phase duration distributions were best explained by an “Erlang” model that requires that each cell cycle phase is viewed as an independently regulated series of limiting steps that must be completed in order to progress to the next phase. The authors propose a “many-for-all” solution to best reconcile the lack of coupling with the heritability of durations between sisters: multiple heritable factors can regulate more than one phase and are propagated independently to daughter cells.
A corollary of such a model is that inducing a dominant effect through a single key regulator of multiple phases would force phase correlations: CDK2 inhibition increased the duration of G1 and S, but also induced coupling between each phase duration. In a second example, DNA damage forced G1-S duration coupling in a U2OS cancer cell line with a defective G1/S checkpoint.
Why we chose this paper
This preprint highlights the value of making simple yet careful quantitative measurements- in more than one cell line! Consistent results across stem cells, non-transformed cell lines and cancer cells considerably strengthen the conclusions of the paper.
These conclusions provide a single conceptual framework that can encompass the lack of coupling between cell cycle phases in steady state, but also allow for genuine differences between cell types (the short G1 of stem cells, for example), as well as enforced coupling from the perturbation of one or a small number of multi-phase regulators. Importantly, the data in this paper suggest that the G1/S and G2/M transitions erase all memory of previous cell cycle phases- if the checkpoints are functioning normally.
In the longer term, the authors’ framework could provide a powerful and generalizable tool to interpret not only dysregulated cell cycles, but also differences in cell cycle architecture between species and cell types.
What next?
Here are a few questions we had after reading the preprint!
- Why does mitosis not fit an Erlang distribution (as one would expect it could also be effectively described as a series of limiting steps)?
- Why is S phase very different in RPE cells compared to H9 or U2OS?
- Why is there so little variability in G1 in stem cells? Is it related to the lineage priming that is thought to happen in G1? Is lineage priming impaired if G1 duration is modified in H9?
- These were cell lines in culture. Could tissue-level interactions apply a degree of external coupling?
- What role does G0 or quiescence play in cell cycle memory (or lack thereof)?
References:
- Spencer, S. L. et al. The Proliferation-Quiescence Decision Is Controlled by a Bifurcation in CDK2 Activity at Mitotic Exit. Cell 155, 369–383 (2013).
- Sakaue-Sawano, A. et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–98 (2008).
- Zetterberg, A. & Larsson, O. Kinetic analysis of regulatory events in G1 leading to proliferation or quiescence of Swiss 3T3 cells. Proc. Natl. Acad. Sci. U. S. A. 82, 5365–5369 (1985).
- Dowling, M. R. et al. Stretched cell cycle model for proliferating lymphocytes. Proc. Natl. Acad. Sci. 111, 6377–6382 (2014).
- Araujo, A. R., Gelens, L., Sheriff, R. S. M. & Santos, S. D. M. Positive Feedback Keeps Duration of Mitosis Temporally Insulated from Upstream Cell-Cycle Events. Mol. Cell 64, 362–375 (2016).
- Sandler, O. et al. Lineage correlations of single cell division time as a probe of cell-cycle dynamics. Nature 519, 468–471 (2015).











 (1 votes)
(1 votes)