Tasting the differences: microbiota analysis of different insect-based novel food
Posted on: 10 May 2020
Preprint posted on 20 February 2020
Article now published in Food Research International at http://dx.doi.org/10.1016/j.foodres.2020.109426
A step forwards for edible insect food safety: core microbiota of processed cricket and mealworm flours, pastas, crackers, and protein bars identified using high throughput DNA sequencing.
Selected by Matt MuzzattiCategories: microbiology
Background
Insects are the potential super food of the future – they are rich in macronutrients, have comparably higher protein content than conventional meat products, and farming insects is more environmentally friendly compared to other livestock. Insects could be a sustainable solution to the problem of food security, but there remain hurdles to gain acceptance among consumers and governments. Food safety is a key component to the worldwide acceptance of edible insects, and safety regulations vary from country to country.
A negative stigma is associated with edible insects: those who do not support the entomophagy movement perceive eating insects as disgusting. Now imagine if a disease outbreak or poisoning occurred because of a lack of research or regulations? It would be a massive blow to an emerging industry seeking to establish insects as a safe and nutritious food source. It’s clear that to successfully promote the entomophagy industry, potential safety risks need to be identified.
Harmful insect microbes represent one of these safety risks. A growing body of literature suggests that growth substrates, diet, insect social behavior, and variable industrial practices can affect the microbial quality of insect food products. This microbial community can even be used as a tracing tool to identify the geographical origin of a food product (Mezzasalma et al. 2017). Using high-throughput DNA sequencing, a high amount of bacterial diversity and variability among whole dried house crickets (Acheta domesticus) and whole dried mealworms (Tenebrio molitor) was identified by Garofalo et al. (2017), and they proposed that some of the microbes they identified COULD act as opportunistic pathogens, such as Listeria spp., Staphylococcus spp., Clostribidum spp., and Bacillus spp.
In this preprint, Frigerio et al. tested the efficacy of high-throughput DNA sequencing to analyze the microbiota variability of 12 commercial edible insect products from 6 different companies in Europe. They examined 4 insect flours, 3 pastas, 2 crackers, and 3 protein bars. Each product only contained 1 of 3 species of insect: house crickets, mealworms or lesser mealworms (Alphitobius diaperinus). They obtained genomic DNA from 250 mg of each food sample and used principal coordinates plots to explore the structure of microbial communities. Heatmap visualization was used to explore microbial abundance.
Key Findings
High-throughput DNA sequencing was effective at analyzing microbial variability among insect food products. All samples yielded good DNA quality, and flour samples had a significantly different microbial community compared to crackers, pasta, and protein bars.
In total, 67 families of bacterial phyla were identified. There was a significant difference among samples belonging to different insects. In fact, each insect had a ‘core microbiota’ formed by a small number of bacteria, although some bacteria were shared between species (Figure 1). These specific microbial signatures could potentially be used as biomarkers to trace insect ingredient origins in food products.
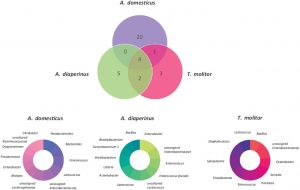
Figure 1. Venn diagram and donut charts of Acheta domesticus, Alphitobius diaperinus and Tenebrio molitor core microbial composition analyzed from 12 different commercial edible insect products including flours, pastas, crackers, and protein bars. From Frigerio et al. 2020 under a CC-BY-NC-ND 4.0 International License.
What I liked about this preprint
The usefulness of high-throughput DNA sequencing at detecting the microbial signatures of edible insect products is a great step for the entomophagy industry. It is a quality assurance protocol that could be standardized and implemented in edible insect regulations to assure food safety.
I found it really cool that each species had a core microbiota associated with it. This should help to avoid and prevent the mislabelling and potential health concerns of insect food products and can hold companies accountable for advertising what species are included in their products. The fish industry has been victim to mislabelling mishaps in the past (Staffen et al. 2017; Xiong et al. 2019), and the entomophagy industry would do well to learn from these mistakes.
Questions for the authors
I assume that all food products have some level of microbial community, so how do the communities you discovered in your research compare with those of pastas, crackers, flours, and protein bars that do not contain any insect product?
You mention that it is conceivable that you only found DNA and no viable cells in the processed food you tested. How would investigations into prebiotic effects help to understand this better? What could be done to test for viable cells in processed edible insect products?
References
Garofalo C, Osimani A, Milanović V, Taccari M, Cardinali F, Aquilanti L, Riolo P, Ruschioni S, Isidoro N, Clementi F (2017) The microbiota of marketed processed edible insects as revealed by high-throughput sequencing. Food Microbiol 62: 15-22. https://doi.org/10.1016/j.fm.2016.09.012
Mezzasalma V, Sandionigi A, Bruni I, Bruno A, Lovicu G, Casiraghi M, Labra M (2017) Grape microbiome as a reliable and persistent signature of field origin and environmental conditions in Cannonau wine production PloS ONE 12(9): e0184615. https://doi.org/10.1371/journal.pone.0184615
Staffen CF, Staffen MD, Becker ML, Löfgren SE, Muniz YCN, de Freitas R, Marrero AR (2017) DNA barcoding reveals the mislabeling of fish in a popular tourist destination in Brazil PeerJ 5: e4006. 10.7717/peerj.4006
Xiong X, Yuan F, Huang M, Lu L, Xiong X, Wen J (2019) DNA barcoding revealed mislabeling and potential health concerns with roasted fish products sold across China. J Food Prot 82(7): 1200-1209.
doi: https://doi.org/10.1242/prelights.20366
Read preprintSign up to customise the site to your preferences and to receive alerts
Register hereAlso in the microbiology category:
Cryo-EM reveals multiple mechanisms of ribosome inhibition by doxycycline
Leonie Brüne
Microbial Feast or Famine: dietary carbohydrate composition and gut microbiota metabolic function
Jasmine Talevi
Citrobacter rodentium infection activates colonic lamina propria group 2 innate lymphoid cells
André Luiz Amorim Costa, Marcus Oliveira
preLists in the microbiology category:
SciELO preprints – From 2025 onwards
SciELO has become a cornerstone of open, multilingual scholarly communication across Latin America. Its preprint server, SciELO preprints, is expanding the global reach of preprinted research from the region (for more information, see our interview with Carolina Tanigushi). This preList brings together biological, English language SciELO preprints to help readers discover emerging work from the Global South. By highlighting these preprints in one place, we aim to support visibility, encourage early feedback, and showcase the vibrant research communities contributing to SciELO’s open science ecosystem.
| List by | Carolina Tanigushi |
BioMalPar XVI: Biology and Pathology of the Malaria Parasite
[under construction] Preprints presented at the (fully virtual) EMBL BioMalPar XVI, 17-18 May 2020 #emblmalaria
| List by | Dey Lab, Samantha Seah |
1
ECFG15 – Fungal biology
Preprints presented at 15th European Conference on Fungal Genetics 17-20 February 2020 Rome
| List by | Hiral Shah |
EMBL Seeing is Believing – Imaging the Molecular Processes of Life
Preprints discussed at the 2019 edition of Seeing is Believing, at EMBL Heidelberg from the 9th-12th October 2019
| List by | Dey Lab |
Antimicrobials: Discovery, clinical use, and development of resistance
Preprints that describe the discovery of new antimicrobials and any improvements made regarding their clinical use. Includes preprints that detail the factors affecting antimicrobial selection and the development of antimicrobial resistance.
| List by | Zhang-He Goh |











 (1 votes)
(1 votes)